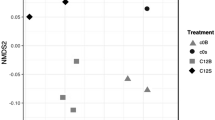
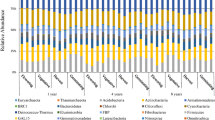
Abstract
Background and aims
Plant-bacterial interactions in the rhizosphere are important in mediating soil nutrient transformations. Plants supply carbon-rich substrates to rhizobacteria as root exudates and bacteria mobilise soil-bound phosphate for plant nutrition. This study aimed to probe the specificity of the plant effect on bacterial gene expression in P-starved rhizosphere conditions.
Methods
DNA microarrays were employed to study gene expression in the rhizosphere of Lolium perenne grown under high and low phosphate regimes (330 μM vs. 3–6 μM phosphate). Root exudation under these regimes was also quantified. Phosphate-regulated gene expression of a panel of 22 genes was compared in rhizosphere, planktonic culture and during biofilm growth on an artificial root.
Results
Plant growth and root exudation were affected by P-availability. P-limited conditions induced increased expression of bacterial genes of an aromatic degradation pathway (catA), heavy metal sensing (PA2523), and membrane proteins (glpM, crcB), while genes involved in cell motility and amino acid uptake/ metabolism were downregulated. A crcB mutant was impaired in rhizosphere survival under low phosphate conditions, though glpM and catA mutants were not affected. Several of the genes studied were induced by phosphate limitation in all three lifestyles studied.
Conclusions
Our results show the importance of the plant-microbe interaction in controlling the bacterial transcriptional response in a phosphate-limited rhizosphere.



Similar content being viewed by others
References
Amikam D, Galperin MY (2006) PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6
Ampe F, Kiss E, Sabourdy F, Batut J (2003) Transcriptome analysis of Sinorhizobium meliloti during symbiosis. Genome Biol 4:R15
Arai H, Mizutani M, Igarashi Y (2003) Transcriptional regulation of the nos genes for nitrous oxide reductase in Pseudomonas aeruginosa. Microbiology 149:29–36
Arasimowicz M, Floryszak-Wieczorek J (2007) Nitric oxide as a bioactive signalling molecule in plant stress responses. Plant Sci 172:876–887
Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM (2006) The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57:233–266
Banin E, Vasil ML, Greenberg EP (2005) Iron and Pseudomonas aeruginosa biofilm formation. Proc Natl Acad Sci USA 102:11076–11081
Barraud N, Hassett DJ, Hwang SH, Rice SA, Kjelleberg S, Webb JS (2006) Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol 188:7344–7353
Barret M, Frey-Klett P, Guillerm-Erckelboudt AY, Boutin M, Guernec G, Sarniguet A (2009) Effect of wheat roots infected with the pathogenic fungus Gaeumannomyces graminis var. tritici on gene expression of the biocontrol bacterium Pseudomonas fluorescens Pf29Arp. Mol Plant-Microbe Interact 22:1611–1623
Barret M, Morrissey JP, O’Gara F (2011) Functional genomics analysis of plant growth-promoting rhizobacterial traits involved in rhizosphere competence. Biol Fertil Soils 47:729–743
Becker A, Berges H, Krol E, Bruand C, Ruberg S, Capela D, Lauber E, Meilhoc E, Ampe F, de Bruijn FJ, Fourment J, Francez-Charlot A, Kahn D, Kuster H, Liebe C, Puhler A, Weidner S, Batut J (2004) Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol Plant-Microbe Interact 17:292–303
Bernard CS, Bordi C, Termine E, Filloux A, de Bentzmann S (2009) Organization and PprB-dependent control of the Pseudomonas aeruginosa tad locus, involved in Flp pilus biology. J Bacteriol 191:1961–1973
Bohm M, Hurek T, Reinhold-Hurek B (2007) Twitching motility is essential for endophytic rice colonization by the N2-fixing endophyte Azoarcus sp. strain BH72. Mol Plant-Microbe Inter 20:526–533
Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193
Buchan A, Crombie B, Alexandre GM (2011) Temporal dynamics and genetic diversity of chemotactic-competent microbial populations in the rhizosphere. Environ Microbiol Rep 12:3171–3184
Byrne SL, Foito A, Hedley PE, Morris JA, Stewart D, Barth S (2011) Early response mechanisms of perennial ryegrass (Lolium perenne) to phosphorus deficiency. Ann Bot 107:243–254
Chatterjee A, Cui Y, Hasegawa H, Chatterjee AK (2007) PsrA, the Pseudomonas sigma regulator, controls regulators of epiphytic fitness, quorum-sensing signals, and plant interactions in Pseudomonas syringae pv. tomato strain DC3000. Appl Environ Microbiol 73:3684–3694
Cornelis P (2008) The ‘core’ and ‘accessory’ regulons of Pseudomonas-specific extracytoplasmic sigma factors. Mol Microbiol 68:810–812
Dakora FD, Phillips DA (2002) Root exudates as mediators of mineral acquisition in low-nutrient environments. Plant Soil 245:35–47
Danhorn T, Fuqua C (2007) Biofilm formation by plant-associated bacteria. Annu Rev Microbiol 61:401–422
Danhorn T, Hentzer M, Givskov M, Parsek MR, Fuqua C (2004) Phosphorus limitation enhances biofilm formation of the plant pathogen Agrobacterium tumefaciens through the PhoR-PhoB regulatory system. J Bacteriol 186:4492–4501
Davey ME, Caiazza NC, O’Toole GA (2003) Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J Bacteriol 185:1027–1036
Deziel E, Lepine F, Milot S, Villemur R (2003) rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149:2005–2013
Dong YH, Zhang XF, Soo HM, Greenberg EP, Zhang LH (2005) The two-component response regulator PprB modulates quorum-sensing signal production and global gene expression in Pseudomonas aeruginosa. Mol Microbiol 56:1287–1301
Dorr J, Hurek T, Reinhold-Hurek B (1998) Type IV pili are involved in plant-microbe and fungus-microbe interactions. Mol Microbiol 30:7–17
Feldman AL, Costouros NG, Wang E, Qian M, Marincola FM, Alexander HR, Libutti SK (2002) Advantages of mRNA amplification for microarray analysis. Biotechniques 33:906
Ferguson GP, Totemeyer S, MacLean MJ, Booth IR (1998) Methylglyoxal production in bacteria: suicide or survival? Arch Microbiol 170:209–219
Fraud S, Campigotto AJ, Chen Z, Poole K (2008) MexCD-OprJ multidrug efflux system of Pseudomonas aeruginosa: involvement in chlorhexidine resistance and induction by membrane-damaging agents dependent upon the AlgU stress response sigma factor. Antimicrob Agents Chemother 52:4478–4482
Gao HC, Yang ZMK, Gentry TJ, Wu LY, Schadt CW, Zhou JZ (2007) Microarray-based analysis of microbial community RNAs by whole-community RNA amplification. Appl Environ Microbiol 73:563–571
Gordon SA, Weber RP (1951) Colorimetric estimation of indoleacetic acid. Plant Physiol 26:192–195
Guvener ZT, Harwood CS (2007) Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol 66:1459–1473
Haddad A, Jensen V, Becker T, Häussler S (2009) The Pho regulon influences biofilm formation and type three secretion in Pseudomonas aeruginosa. Environ Microbiol Rep 1:488–494
Hentzer M, Teitzel GM, Balzer GJ, Heydorn A, Molin S, Givskov M, Parsek MR (2001) Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol 183:5395–5401
Hershberger CD, Ye RW, Parsek MR, Xie ZD, Chakrabarty AM (1995) The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative σ factor (σE). Proc Natl Acad Sci USA 92:7941–7945
Hinsinger P (2001) Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: a review. Plant Soil 237:173–195
Hinsinger P, Gobran GR, Gregory PJ, Wenzel WW (2005) Rhizosphere geometry and heterogeneity arising from root-mediated physical and chemical processes. New Phytol 168:293–303
Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP (1998) A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86
Hsieh YJ, Wanner BL (2010) Global regulation by the seven-component Pi signaling system. Curr Opin Microbiol 13:198–203
Huang CY, Roessner U, Eickmeier I, Genc Y, Callahan DL, Shirley N, Langridge P, Bacic A (2008) Metabolite profiling reveals distinct changes in carbon and nitrogen metabolism in phosphate-deficient barley plants (Hordeum vulgare L.). Plant Cell Physiol 49:691–703
Inoue T, Shingaki R, Hirose S, Waki K, Mori H, Fukui K (2007) Genome-wide screening of genes required for swarming motility in Escherichia coli K-12. J Bacteriol 189:950–957
Iswandi A, Bossier P, Vandenabeele J, Verstraete W (1987) Influence of the inoculation density of the rhizopseudomonad strain 7NSK2 on the growth and the composition of the root microbial community of maize (Zea mays) and barley (Hordeum vulgare). Biol Fertil Soils 4:119–123
Jenal U, Malone J (2006) Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet 40:385–407
Jensen V, Lons D, Zaoui C, Bredenbruch F, Meissner A, Dieterich G, Munch R, Haussler S (2006) RhlR expression in Pseudomonas aeruginosa is modulated by the Pseudomonas quinolone signal via PhoB-dependent and -independent pathways. J Bacteriol 188:8601–8606
Ju HS, Yokoigawa K, Misono H, Ohnishi K (2005) Cloning of alanine racemase genes from Pseudomonas fluorescens strains and oligomerization states of gene products expressed in Escherichia coli. J Biosci Bioeng 100:409–417
Karunakaran R, Ramachandran VK, Seaman JC, East AK, Mouhsine B, Mauchline TH, Prell J, Skeffington A, Poole PS (2009) Transcriptomic analysis of Rhizobium leguminosarum biovar viciae in symbiosis with host plants Pisum sativum and Vicia cracca. J Bacteriol 191:4002–4014
Korbsrisate S, Vanaporn M, Kerdsuk P, Kespichayawattana W, Vattanaviboon P, Kiatpapan P, Lertmemongkolchai G (2005) The Burkholderia pseudomallei RpoE (AlgU) operon is involved in environmental stress tolerance and biofilm formation. FEMS Microbiol Lett 252:243–249
Lambers H, Shane MW, Cramer MD, Pearse SJ, Veneklaas EJ (2006) Root structure and functioning for efficient acquisition of phosphorus: matching morphological and physiological traits. Ann Bot 98:693–713
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodefellow M (eds) Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom, pp 115–175
Lee SW, Edlin G (1985) Expression of tetracycline resistance in pBR322 derivatives reduces the reproductive fitness of plasmid-containing Escherichia coli. Gene 39:173–180
Leveau JH, Lindow SE (2005) Utilization of the plant hormone indole-3-acetic acid for growth by Pseudomonas putida strain 1290. Appl Environ Microbiol 71:2365–2371
Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98:31–36
Madhusudhan KT, Lorenz D, Sokatch JR (1993) The bkdR gene of Pseudomonas putida is required for expression of the bkd operon and encodes a protein related to Lrp of Escherichia coli. J Bacteriol 175:3934–3940
Marilley L, Aragno M (1999) Phylogenetic diversity of bacterial communities differing in degree of proximity of Lolium perenne and Trifolium repens roots. Appl Soil Ecol 13:127–136
Mark GL, Dow JM, Kiely PD, Higgins H, Haynes J, Baysse C, Abbas A, Foley T, Franks A, Morrissey J, O’Gara F (2005) Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe-plant interactions. Proc Natl Acad Sci USA 102:17454–17459
Marschner P, Solaiman Z, Rengel Z (2006) Rhizosphere properties of Poaceae genotypes under P-limiting conditions. Plant Soil 283:11–24
Matilla MA, Espinosa-Urgel M, Rodriguez-Herva JJ, Ramos JL, Ramos-Gonzalez MI (2007) Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere. Genome Biol 8:R179
Mattick JS (2002) Type IV pili and twitching motility. Annu Rev Microbiol 56:289–314
McPhee JB, Lewenza S, Hancock RE (2003) Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol 50:205–217
Meharg AA, Killham K (1995) Loss of exudates from the roots of perennial ryegrass inoculated with a range of microorganisms. Plant Soil 170:345–349
Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S (2007) The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol 65:876–895
Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, Doumas P, Nacry P, Herrerra-Estrella L, Nussaume L, Thibaud MC (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102:11934–11939
Monds RD, Silby MW, Mahanty HK (2001) Expression of the Pho regulon negatively regulates biofilm formation by Pseudomonas aureofaciens PA147-2. Mol Microbiol 42:415–426
Monds RD, Newell PD, Gross RH, O’Toole GA (2007) Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol Microbiol 63:656–679
Morcuende R, Bari R, Gibon Y, Zheng WM, Pant BD, Blasing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, Scheible WR (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30:85–112
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Nakada Y, Itoh Y (2002) Characterization and regulation of the gbuA gene, encoding guanidinobutyrase in the arginine dehydrogenase pathway of Pseudomonas aeruginosa PAO1. J Bacteriol 184:3377–3384
O’Toole GA, Gibbs KA, Hager PW, Phibbs PV, Kolter R (2000) The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J Bacteriol 182:425–431
Ochsner UA, Wilderman PJ, Vasil AI, Vasil ML (2002) GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol Microbiol 45:1277–1287
Palmer BR, Marinus MG (1994) The dam and dcm strains of Escherichia coli—A review. Gene 143:1–12
Pamp SJ, Tolker-Nielsen T (2007) Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J Bacteriol 189:2531–2539
Paterson E, Sim A (1999) Rhizodeposition and C-partitioning of Lolium perenne in axenic culture affected by nitrogen supply and defoliation. Plant Soil 216:155–164
Paterson E, Gebbing T, Abel C, Sim A, Telfer G (2007) Rhizodeposition shapes rhizosphere microbial community structure in organic soil. New Phytol 173:600–610
Pearse SJ, Veneklaas EJ, Cawthray GR, Bolland MDA, Lambers H (2006) Carboxylate release of wheat, canola and 11 grain legume species as affected by phosphorus status. Plant Soil 288:127–139
Pflock M, Finsterer N, Joseph B, Mollenkop H, Meyer TF, Beier D (2006) Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. J Bacteriol 188:3449–3462
Poole K (2001) Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J Mol Microbiol Biotechnol 3:255–264
Reva ON, Weinel C, Weinel M, Bohm K, Stjepandic D, Hoheisel JD, Tummler B (2006) Functional genomics of stress response in Pseudomonas putida KT2440. J Bacteriol 188:4079–4092
Richardson AE (2001) Prospects for using soil microorganisms to improve the acquisition of phosphorus by plants. Aust J Plant Physiol 28:897–906
Richardson AE, Hocking PJ, Simpson RJ, George TS (2009) Plant mechanisms to optimise access to soil phosphorus. Crop Pasture Sci 60:124–143
Römling U, Gomelsky M, Galperin MY (2005) C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol 57:629–639
Rooney DC, Clipson NJW (2009) Phosphate addition and plant species alters microbial community structure in acidic upland grassland soil. Microb Ecol 57:4–13
Ruffel S, Freixes S, Balzergue S, Tillard P, Jeudy C, Martin-Magniette ML, van der Merwe MJ, Kakar K, Gouzy J, Fernie AR, Udvardi M, Salon C, Gojon A, Lepetit M (2008) Systemic signaling of the plant nitrogen status triggers specific transcriptome responses depending on the nitrogen source in Medicago truncatula. Plant Physiol 146:2020–2035
Sanchez-Calderon L, Lopez-Bucio J, Chacon-Lopez A, Cruz-Ramirez A, Nieto-Jacobo F, Dubrovsky JG, Herrera-Estrella L (2005) Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant Cell Physiol 46:174–184
Sand O, Gingras M, Beck N, Hall C, Trun N (2003) Phenotypic characterization of overexpression or deletion of the Escherichia coli crcA, cspE and crcB genes. Microbiology 149:2107–2117
Schenk A, Weingart H, Ullrich MS (2008) The alternative sigma factor AlgT, but not alginate synthesis, promotes in planta multiplication of Pseudomonas syringae pv. glycinea. Microbiology 154:413–421
Semmler AB, Whitchurch CB, Leech AJ, Mattick JS (2000) Identification of a novel gene, fimV, involved in twitching motility in Pseudomonas aeruginosa. Microbiology 146(Pt 6):1321–1332
Shaw LJ, Morris P, Hooker JE (2006) Perception and modification of plant flavonoid signals by rhizosphere microorganisms. Environ Microbiol 8:1867–1880
Smyth GK (2004) Linear models and empirical Bayes for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3
Smyth GK (2005) Limma: Linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W (eds) Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer, New York, pp 397–420
Stanley NR, Lazazzera BA (2004) Environmental signals and regulatory pathways that influence biofilm formation. Mol Microbiol 52:917–924
Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100:9440–9445
Strych U, Huang HC, Krause KL, Benedik MJ (2000) Characterization of the alanine racemases from Pseudomonas aeruginosa PAO1. Curr Microbiol 41:290–294
Weger LAD, Dekkers LC, Vanderbij AJ, Lugtenberg BJJ (1994) Use of phosphate-reporter bacteria to study phosphate limitation in the rhizosphere and in bulk soil. Molec Plant-Microbe Interactions 7:32–38
Weinberg Z, Wang J, Bogue J, Yang J, Corbino K, Moy R, Breaker R (2010) Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol 11:R31
Weinrick B, Dunman PM, McAleese F, Murphy E, Projan SJ, Fang Y, Novick RP (2004) Effect of mild acid on gene expression in Staphylococcus aureus. J Bacteriol 186:8407–8423
Wozniak DJ, Wyckoff TJO, Starkey M, Keyser R, Azadi P, O’Toole GA, Parsek MR (2003) Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc Natl Acad Sci USA 100:7907–7912
Yuan ZC, Zaheer R, Morton R, Finan TM (2006) Genome prediction of PhoB-regulated promoters in Sinorhizobium meliloti and twelve proteobacteria. Nucleic Acids Res 34:2686–2697
Acknowledgements
This work was supported by the Natural Environment Research Council (NERC) and by a CASE studentship to AZ sponsored by the Biotechnology and Biological Sciences Research Council (BBSRC) and Macaulay Enterprises Ltd.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Jorge Vivanco.
Rights and permissions
About this article
Cite this article
Zyśko, A., Sanguin, H., Hayes, A. et al. Transcriptional response of Pseudomonas aeruginosa to a phosphate-deficient Lolium perenne rhizosphere. Plant Soil 359, 25–44 (2012). https://doi.org/10.1007/s11104-011-1060-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-011-1060-z