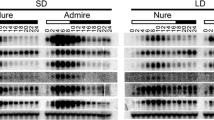
Abstract
A family of CBF transcription factors plays a major role in reconfiguring the plant transcriptome in response to low-freezing temperature in temperate cereals. In barley, more than 13 HvCBF genes map coincident with the major QTL FR-H2 suggesting them as candidates to explain the function of the locus. Variation in copy number (CNV) of specific HvCBFs was assayed in a panel of 41 barley genotypes using RT-qPCR. Taking advantage of an accurate phenotyping that combined Fv/Fm and field survival, resistance-associated variants within FR-H2 were identified. Genotypes with an increased copy number of HvCBF4 and HvCBF2 (at least ten and eight copies, respectively) showed greater frost resistance. A CAPS marker able to distinguish the CBF2A, CBF2B and CBF2A/B forms was developed and showed that all the higher-ranking genotypes in term of resistance harbour only CBF2A, while other resistant winter genotypes harbour also CBF2B, although at a lower CNV. In addition to the major involvement of the HvCBF4-HvCBF2 genomic segment in the proximal cluster of CBF elements, a negative role of HvCBF3 in the distal cluster was identified. Multiple linear regression models taking into account allelic variation at FR-H1/VRN-H1 explained 0.434 and 0.550 (both at p < 0.001) of the phenotypic variation for Fv/Fm and field survival respectively, while no interaction effect between CNV at the HvCBFs and FR-H1/VRN-H1 was found. Altogether our data suggest a major involvement of the CBF genes located in the proximal cluster, with no apparent involvement of the central cluster contrary to what was reported for wheat.




Similar content being viewed by others
Abbreviations
- FR :
-
Frost resistance loci
- CBF/DREB:
-
C-Repeat binding factor/dehydration responsive element binding factor proteins
- CRT/DRE:
-
C-RepeaT/dehydration responsive elements
- VRN :
-
Vernalization requirement genes and loci
- CNV:
-
Copy number variation
- CNVs:
-
Copy number variants
References
Akar T, Francia E, Tondelli A, Rizza F, Stanca AM, Pecchioni N (2009) Marker-assisted characterization of frost tolerance in barley (Hordeum vulgare L.) Plant Breed 128:381–386. doi:10.1111/j.1439-0523.2008.01553.x
Alkan C, Coe BP, Eichler EE (2011) Genome structural variation discovery and genotyping. Nat Rev Genet 12:363–376. doi:10.1038/nrg2958
Badawi M, Danyluk J, Boucho B, Houde M, Sarhan F (2007) The CBF gene family in hexaploid wheat and its relationship to the phylogenetic complexity of cereal CBFs. Mol Genet Genomics 277:533–554. doi:10.1007/s00438-006-0206-9
Campoli C, Matus-Cádiz MA, Pozniak CJ, Cattivelli L, Fowler DB (2009) Comparative expression of Cbf genes in the Triticeae under different acclimation induction temperatures. Mol Genet Genomics 282:141–152. doi:10.1007/s00438-009-0451-9
Cook DE, Lee TG, Guo X, Melito S, Wang K, Bayless AM, Wang J, Hughes TJ, Willis DK, Clemente TE, Diers BW, Jiang J, Hudson ME, Bent AF (2012) Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338:1206–1209. doi:10.1126/science.1228746
D’haene B, Vandesompele J, Hellemans J (2010) Accurate and objective copy number profiling using real-time quantitative PCR. Methods 50:262–270. doi:10.1016/j.ymeth.2009.12.007
Deng W, Casao MC, Wang P, Sato K, Hayes PM, Finnegan EJ, Trevaskis B (2015) Direct links between the vernalization response and other key traits of cereal crops. Nat Commun 6:5882. doi:10.1038/ncomms6882
Dhillon T, Stockinger EJ (2013) Cbf14 copy number variation in the A, B, and D genomes of diploid and polyploid wheat. Theor Appl Genet 126:2777–2789. doi:10.1007/s00122-013-2171-0
Dhillon T, Pearce SP, Stockinger EJ, Distelfeld A, Li C, Knox AK, Vashegyi I, Galiba G, Vágú A, Galiba G, Dubcovsky J (2010) Regulation of freezing tolerance and flowering in temperate cereals: The VRN-1 connection. Plant Physiol 153:1846–1858. doi:10.1104/pp.110.159079
Francia E, Rizza F, Cattivelli L, Stanca AM, Galiba G, Tóth B, Hayes PM, Skinner JS, Pecchioni N (2004) Two loci on chromosome 5 H determine low-temperature tolerance in a “Nure” (winter) × “Tremois” (spring) barley map. Theor Appl Genet 108:670–680. doi:10.1007/s00122-003-1468-9
Francia E, Barabaschi D, Tondelli A, Laidò G, Rizza F, Stanca AM, Busconi M, Fogher C, Stockinger EJ, Pecchioni N (2007) Fine mapping of a HvCBF gene cluster at the frost resistance locus Fr-H2 in barley. Theor Appl Genet 115:1083–1091. doi:10.1007/s00122-007-0634-x
Francia E, Pecchioni N, Policriti A, Scalabrin S (2015) CNV and sStructural variation in plants: prospects of NGS approaches. In: Sablok G, Kumar S, Ueno S, Kuo J, Varotto C (eds) Advances in the understanding of biological sciences using next generation sequencing (NGS) approaches. Springer Cham, pp 211–232. doi:10.1007/978-3-319-17157-9
Galiba G, Stockinger EJ, Francia E, Milc J, Kocsy G, Pecchioni N (2013) Freezing tolerance in the Triticeae. In: Varshney RK, Tuberosa R (eds) Translational genomics for crop breeding. Wiley, New york, pp 99–124. doi:10.1002/9781118728475
International Barley Genome Sequencing Consortium, Mayer KFX, Waugh R, Brown JWS, Schulman A, Langridge P, Platzer M, Fincher GB, Muehlbauer GJ, Sato K, Close TJ, Wise RP, Stein N (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491:711–716. doi:10.1038/nature11543
Jeknić Z, Pillman KA, Dhillon T, Skinner JS, Veisz O, Cuesta-Marcos A, Hayes PM, Jacobs AK, Chen THH, Stockinger EJ (2014) Hv-CBF2A overexpression in barley accelerates COR gene transcript accumulation and acquisition of freezing tolerance during cold acclimation. Plant Mol Biol 84:67–82. doi:10.1007/s11103-013-0119-z
Jiao Y, Zhao H, Ren L, Song W, Zeng B, Guo J, Wang B, Liu Z, Chen J, Li W, Zhang M, Xie S, Lai J (2012) Genome-wide genetic changes during modern breeding of maize. Nat Genet 44:812–815. doi:10.1038/ng.2312
Knox AK, Li C, Vágújfalvi A, Galiba G, Stockinger EJ, Dubcovsky J (2008) Identification of candidate CBF genes for the frost tolerance locus Fr-A m 2 in Triticum monococcum. Plant Mol Biol 67:257–270. doi:10.1007/s11103-008-9316-6
Knox AK, Dhillon T, Cheng H, Tondelli A, Pecchioni N, Stockinger EJ (2010) CBF gene copy number variation at Frost Resistance-2 is associated with levels of freezing tolerance in temperate-climate cereals. Theor Appl Genet 121:21–35. doi:10.1007/s00122-010-1288-7
Kosová K, Prášil IT, Vítámvás P (2008) The relationship between vernalization- and photoperiodically-regulated genes and the development of frost tolerance in wheat and barley. Biol Plant 52:601–615. doi:10.1007/s10535-008-0120-6
Levitt J (1980) Chilling, freezing, and high temperature stresses. In: Levitt J (ed) In: response of plants to environmental stresses, 2nd edn. vol 1. Academic Press Inc., New York, NY
Li W, Olivier M (2013) Current analysis platforms and methods for detecting copy number variation. Physiol Genomics 45:1–16. doi:10.1152/physiolgenomics.00082.2012
Limin AE, Fowler DB (2006) Low-temperature tolerance and genetic potential in wheat (Triticum aestivum L.): response to photoperiod, vernalization, and plant development. Planta 224:360–366. doi:10.1007/s00425-006-0219-y
Ma L, Chung WK (2014) Quantitative analysis of copy number variants based on real-time LightCycler PCR. Curr Protoc Hum Genet 80:7.21.1–7.21.8. doi:10.1002/0471142905.hg0721s80
Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TFC, McCarroll SA, Visscher PM (2009) Finding the missing heritability of complex diseases. Nature 461:747–753. doi:10.1038/nature08494
Maron LG, Guimarães CT, Kirst M, Albert PS, Birchler JA, Bradbury PJ, Buckler ES, Coluccio AE, Danilova TV, Kudrna D, Magalhaes JV, Piñeros MA, Schatz MC, Wing RA, Kochian LV (2013) Aluminum tolerance in maize is associated with higher MATE1 gene copy number. Proc Natl Acad Sci USA 110:5241–5246. doi:10.1073/pnas.1220766110
Marozsán-Tóth Z, Vashegyi I, Galiba G, Tóth B (2015) The cold response of CBF genes in barley is regulated by distinct signaling mechanisms. J Plant Physiol 181:42–49. doi:10.1016/j.jplph.2015.04.002
McHale LK, Haun WJ, Xu WW, Bhaskar PB, Anderson JE, Hyten DL, Gerhardt DJ, Jeddeloh JA, Stupar RM (2012) Structural variants in the soybean genome localize to clusters of biotic stress-response genes. Plant Physiol 159:1295–1308. doi:10.1104/pp.112.194605
Mickelbart MV, Hasegawa PM, Bailey-Serres J (2015) Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat Rev Genet 16:237–251. doi:10.1038/nrg3901
Miller AK, Galiba G, Dubcovsky J (2006) A cluster of 11 CBF transcription factors is located at the frost tolerance locus Fr-A m 2 in Triticum monococcum. Mol Genet Genomics 275:193–203. doi:10.1007/s00438-005-0076-6
Mohseni S, Che H, Djillali Z, Dumont E, Nankeu J, Danyluk J (2012) Wheat CBF gene family: identification of polymorphisms in the CBF coding sequence. Genome 55:865–881. doi:10.1139/gen-2012-0112
Morran S, Eini O, Pyvovarenko T, Parent B, Singh R, Ismagul A, Eliby S, Shirley N, Langridge P, Lopato S (2011) Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol J 9:230–249. doi:10.1111/j.1467-7652.2010.00547.x
Muñoz-Amatriaín M, Eichten SR, Wicker T, Richmond TA, Mascher M, Steuernagel B, Scholz U, Ariyadasa R, Spannagl M, Nussbaumer T, Mayer KF, Taudien S, Platzer M, Jeddeloh JA, Springer NM, Muehlbauer GJ, Stein N (2013) Distribution, functional impact, and origin mechanisms of copy number variation in the barley genome. Genome Biol 14:R58. doi:10.1186/gb-2013-14-6-r58
Nitcher R, Distelfeld A, Tan C, Yan L, Dubcovsky J (2013) Increased copy number at the HvFT1 locus is associated with accelerated flowering time in barley. Mol Genet Genomics 288:261–275. doi:10.1007/s00438-013-0746-8
Novillo F, Medina J, Rodríguez-Franco M, Neuhaus G, Salinas J (2011) Genetic analysis reveals a complex regulatory network modulating CBF gene expression and Arabidopsis response to abiotic stress. J Exp Bot 63(1):293–304. doi:10.1093/jxb/err279
Pasquariello M, Barabaschi D, Himmelbach A, Steuernagel B, Ariyadasa R, Stein N, Gandolfi F, Tenedini E, Bernardis I, Tagliafico E, Pecchioni N, Francia E (2014) The barley Frost resistance-H2 locus. Funct Integr Genomics 14(1):85–100. doi:10.1007/s10142-014-0360-9
Payne R (2014) A guide to regression, nonlinear and generalized linear models in GenStat, 17th edn. VSN International, Hemel Hempstead
Pearce S, Zhu J, Boldizsár Á, Vágújfalvi A, Burke A, Garland-Campbell K, Galiba G, Dubcovsky J (2013) Large deletions in the CBF gene cluster at the Fr-B2 locus are associated with reduced frost tolerance in wheat. Theor Appl Genet 126:2683–2697. doi:10.1007/s00122-013-2165-y
Pecchioni N, Kosová K, Vítámvás P, Prášil IT, Milc JA, Francia E, Gulyás Z, Kocsy G, Galiba G (2012) Genomics of low-temperature tolerance for an increased sustainability of wheat and barley production. In: Tuberosa R, Graner A, Frison E (eds) Genomics of plant genetic resources to improve crop production, food security and nutritional quality. Springer, Berlin, pp 149–183. doi:10.1007/978-94-007-7575-6
Poland JA, Brown PJ, Sorrells ME, Jannink J-L (2012) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7(2):e32253. doi:10.1371/journal.pone.0032253
Rizza F, Pagani D, Gut M, Prášil IT, Lago C, Tondelli A, Orrù L, Mazzucotelli E, Francia E, Badeck F-W, Crosatti C, Terzi V, Cattivelli L, Stanca AM (2011) Diversity in the response to low temperature in representative barley genotypes cultivated in Europe. Crop Sci 51:2759–2779. doi:10.2135/cropsci2011.01.0005
Skinner JS, von Zitzewitz J, Szucs P, Marquez-Cedillo L, Filichkin T, Amundsen K, Stockinger EJ, Thomashow MF, Chen THH, Hayes PM (2005) Structural, functional, and phylogenetic characterization of a large CBF gene family in barley. Plant Mol Biol 59:533–551. doi:10.1007/s11103-005-2498-2
Skinner JS, Szucs P, von Zitzewitz J, Marquez-Cedillo L, Filichkin T, Stockinger EJ, Thomashow MF, Chen THH, Hayes PM (2006) Mapping of barley homologs to genes that regulate low temperature tolerance in Arabidopsis. Theor Appl Genet 112:832–842. doi:10.1007/s00122-005-0185-y
Soltész A, Smedley M, Vashegyi I, Galiba G, Harwood W, Vagu jfalvi A (2013) Transgenic barley lines prove the involvement of TaCBF14 and TaCBF15 in the cold acclimation process and in frost tolerance. J Exp Bot 64:1849–1862. doi:10.1093/jxb/ert050
Stockinger EJ, Skinner JS, Gardner KG, Francia E, Pecchioni N (2007) Expression levels of barley Cbf genes at the Frost resistance-H2 locus are dependent upon alleles at Fr-H1 and Fr-H2. Plant J 51:308–321. doi:10.1111/j.1365-313X.2007.0141.x
Sutton T, Baumann U, Hayes J, Collins NC, Shi B-J, Schnurbusch T, Hay A, Mayo G, Pallotta M, Tester M, Langridge P (2007) Boron-toxicity tolerance in barley arising from efflux transporter amplification. Science 318:1446–1449. doi:10.1126/science.1146853
Swanson-Wagner RA, Eichten SR, Kumari S, Tiffin P, Stein JC, Ware D, Springer NM (2010) Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor. Genome Res 20:1689–1699. doi:10.1101/gr.109165.110
Thomashow MF (2010) Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol 154:571–577. doi:10.1104/pp.110.161794
Tondelli A, Francia E, Barabaschi D, Pasquariello M, Pecchioni N (2011) Inside the CBF locus in Poaceae. Plant Sci 180:39–45
Tondelli A, Pagani D, Ghafoori IN, Rahimi M, Ataei R, Rizza F, Flavell AJ, Cattivelli L (2014) Allelic variation at Fr-H1/Vrn-H1 and Fr-H2 loci is the main determinant of frost tolerance in spring barley. Environ Exp Bot 106:148–155. doi:10.1016/j.envexpbot.2014.02.014
Tranquilli G, Dubcovsky J (2000) Epistatic interaction between vernalization genes Vrn-A m 1 and Vrn-A m 2 in diploid wheat. J Hered 91:304–306
Visioni A, Tondelli A, Francia E, Pswarayi A, Malosetti M, Russell J, Thomas W, Waugh R, Pecchioni N, Romagosa I, Comadran J (2013) Genome-wide association mapping of frost tolerance in barley (Hordeum vulgare L.) BMC Genomics 14:424. doi:10.1186/1471-2164-14-424
von Zitzewitz J, Szucs P, Dubcovsky J, Yan L, Francia E, Pecchioni N, Casas A, Chen THH, Hayes PM, Skinner JS (2005) Molecular and structural characterization of barley vernalization genes. Plant Mol Biol 59:449–467. doi:10.1007/s11103-005-0351-2
von Zitzewitz J, Cuesta-Marcos A, Condon F, Castro AJ, Chao S, Corey A, Filichkin T, Fisk SP, Gutierrez L, Haggard K, Karsai I, Muehlbauer GJ, Smith KP, Veisz O, Hayes PM (2011) The genetics of winterhardiness in barley: perspectives from genome-wide association mapping. Plant Genome J 4:76. doi:10.3835/plantgenome2010.12.0030
Weaver S, Dube S, Mir A, Qin J, Sun G, Ramakrishnan R, Jones RC, Livak KJ (2010) Taking qPCR to a higher level: analysis of CNV reveals the power of high throughput qPCR to enhance quantitative resolution. Methods 50:271–276. doi:10.1016/j.ymeth.2010.01.003
Zhu J, Pearce S, Burke A, See DR, Skinner DZ, Dubcovsky J, Garland-Campbell K (2014) Copy number and haplotype variation at the VRN-A1 and central FR-A2 loci are associated with frost tolerance in hexaploid wheat. Theor Appl Genet 127:1183–1197. doi:10.1007/s00122-014-2290-2
Zmieńko A, Samelak A, Kozłowski P, Figlerowicz M (2013) Copy number polymorphism in plant genomes. Theor Appl Genet 127:1–18. doi:10.1007/s00122-013-2177-7
Acknowledgments
This work was partially supported by Fondazione Cassa di Risparmio di Modena, Progetto di Ricerca Internazionale “FROSTMAP—Physical mapping of the barley Frost resistance-H2 (Fr-H2) locus”. Funding from “GENOMORE” project of the Fondazione Cassa di Risparmio di Carpi is also acknowledged. The authors are grateful to Dr. Franz W. Badeck (PIK—Potsdam, Germany) for his critical suggestions during statistical analysis and to Mrs. Donata Pagani (CRA-GPG, Fiorenzuola, Italy) for the excellent technical assistance during phenotypic analysis, and to Dr. Ilja Tom Prášil (Crop Research Institute, Prague, Czech Republic) for providing some barley varieties.
Author contributions
All the authors have participated in the work and none of them have any conflict of interest. In particular, Enrico Francia and Nicola Pecchioni conceived the study. Enrico Francia and Caterina Morcia designed sequence-specific amplicons on the tested genes, carried out RT-qPCR and copy number calculation. Marianna Pasquariello and Valentina Mazzamurro developed a novel CAPS marker and together with Justyna Milc performed CBF2A/B screening. Fulvia Rizza and Valeria Terzi performed the phenotypic characterization for frost resistance of the plant material. Nicola Pecchioni contributed to analysis and interpretation of data and participated in the discussion of results. The manuscript was written Enrico Francia and Justyna Milc.
Author information
Authors and Affiliations
Corresponding author
Additional information
E. Francia and C. Morcia have equally contributed to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2016_505_MOESM2_ESM.tif
Supplementary Fig. 1 Graphical genotyping and frost resistance of ‘Nure’ x ‘Tremois’ F5:6 recombinants in the HvCBF cluster at Fr-H2. a Graphical representation of chromosome 5H region of the phenotyped F5:6 plants. Markers name and order derive from Francia et al. (2007) and Pasquariello et al. (2014). Chromosomal segments with ‘Nure’ and ‘Tremois’ marker alleles are in dark and light gray, respectively. NT-42 and NT-64 are DH lines of the ‘NxT’ population developed by Francia et al. (2004). The number of genotypes considered for each recombinant class is in parenthesis. b Bar graph for Fv/Fm representing the mean ± s.e. for n = 5 independent experiments in controlled conditions (freezing stress ranged from −11°C to −13°C). c Bar graph for winter survival representing the mean ± s.e. for a field trial in 2012/13 (min temp. −7°C). Lowercase letters in (b) and (c) represent least significant differences (LSD0.05) (TIF 1956 KB)
Rights and permissions
About this article
Cite this article
Francia, E., Morcia, C., Pasquariello, M. et al. Copy number variation at the HvCBF4–HvCBF2 genomic segment is a major component of frost resistance in barley. Plant Mol Biol 92, 161–175 (2016). https://doi.org/10.1007/s11103-016-0505-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-016-0505-4