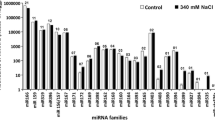
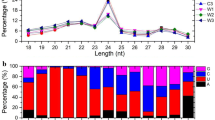
Abstract
Myco-heterotroph Monotropa hypopitys is a widely spread perennial herb used to study symbiotic interactions and physiological mechanisms underlying the development of non-photosynthetic plant. Here, we performed, for the first time, transcriptome-wide characterization of M. hypopitys miRNA profile using high throughput Illumina sequencing. As a result of small RNA library sequencing and bioinformatic analysis, we identified 55 members belonging to 40 families of known miRNAs and 17 putative novel miRNAs unique for M. hypopitys. Computational screening revealed 206 potential mRNA targets for known miRNAs and 31 potential mRNA targets for novel miRNAs. The predicted target genes were described in Gene Ontology terms and were found to be involved in a broad range of metabolic and regulatory pathways. The identification of novel M. hypopitys-specific miRNAs, some with few target genes and low abundances, suggests their recent evolutionary origin and participation in highly specialized regulatory mechanisms fundamental for non-photosynthetic biology of M. hypopitys. This global analysis of miRNAs and their potential targets in M. hypopitys provides a framework for further investigation of miRNA role in the evolution and establishment of non-photosynthetic myco-heterotrophs.



Similar content being viewed by others
References
Abdel-Ghany SE, Pilon M (2008) MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J BiolChem 283(23):15932–15945. doi:10.1074/jbc.M801406200
Alaba S, Piszczalka P, Pietrykowska H, Pacak AM, Sierocka I, Nuc PW, Singh K, Plewka P, Sulkowska A, Jarmolowski A, Karlowski WM, Szweykowska-Kulinska Z (2015) The liverwort Pelliaendiviifolia shares microtranscriptomic traits that are common to green algae and land plants. New Phytol 206(1):352–367. doi:10.1111/nph.13220
Allen E, Xie ZX, Gustafson AM, Sung GH, Spatafora JW, Carrington JC (2004) Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet 36:1282–1290. doi:10.1038/ng1478
Alonso-Peral MM, Sun C, Millar AA (2012) MicroRNA159 Can Act as a Switch or Tuning MicroRNA Independently of Its Abundance in Arabidopsis. PLoS One 7(4):e34751. doi:10.1371/journal.pone.0034751
Arazi T (2012) MicroRNAs in the moss Physcomitrella patens. Plant MolBiol 80(1):55–65. doi:10.1007/s11103-011-9761-5
Axtell MJ, Snyder JA, Bartel DP (2007) Common functions for diverse small RNAs of land plants. Plant Cell 19(6):1750–1769. doi:10.1105/tpc.107.051706
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
Bazin J, Khan GA, Combier JP, Bustos-Sanmamed P, Debernardi JM, Rodriguez R, Sorin C, Palatnik J, Hartmann C, Crespi M, Lelandais-Brière C (2013) miR396 affects mycorrhization and root meristem activity in the legume Medicagotruncatula. Plant J 74(6):920–934. doi:10.1111/tpj.12178
Bell EM, Lin WC, Husbands AY, Yu L, Jaganatha V, Jablonska B, Mangeon A, Neff MM, Girke T, Springer PS (2012) Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc Natl Acad Sci USA 109(51):21146–21151. doi:10.1073/pnas.1210789109
Bhardwaj J, Chauhan R, Swarnkar MK, Chahota RK, Singh AK, Shankar R, Yadav SK (2013) Comprehensive transcriptomic study on horse gram (Macrotylomauniflorum): De novo assembly, functional characterization and comparative analysis in relation to drought stress. BMC Genom 14:647. doi:10.1186/1471-2164-14-647
Bidartondo MI (2005) The evolutionary ecology of myco-heterotrophy. New Phytol 167(2):335–352. doi:10.1111/j.1469-8137.2005.01429.x
Bowman JL (2013) Walk about on the long branches of plant evolution. Curr Opin Plant Biol 16(1):70–77. doi:10.1016/j.pbi.2012.10.001
Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320(5880):1185–1190. doi:10.1126/science.1159151
Budak H, Kantar M, Bulut R, Akpinar BA (2015) Stress responsive miRNAs and isomiRs in cereals. Plant Sci 235:1–13. doi:10.1016/j.plantsci.2015.02.008
Cakir O, Candar-Cakir B, Zhang B (2015) Small RNA and degradome sequencing reveals important microRNA function in Astragaluschrysochlorus response to selenium stimuli. Plant Biotechnol J. doi:10.1111/pbi.12397
Cara B, Giovannoni JJ (2008) Molecular biology of ethylene during tomato fruit development and maturation. Plant Sci 175:106–113. doi:10.1016/j.plantsci.2008.03.021
Chavez Montes RA, Rosas-Cárdenas FF, DePaoli E, Accerbi M, Meyers BC, Rymarquis LA, Mahalingam G, Marsch-Martinez N, Meyers BC, Green PJ, de Folter S (2014) Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nat Commun 5:3722. doi:10.1038/ncomms4722
Chen X (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25:21–44. doi:10.1146/annurev.cellbio.042308.113417
Chen L, Ren Y, Zhang Y, Xu J, Sun F, Zhang Z, Wang Y (2012) Genome-wide identification and expression analysis of heat-responsive and novel microRNAs in Populustomentosa. Gene 504(2):160–165. doi:10.1016/j.gene.2012.05.034
Chen X, Chan WL, Zhu FY, Lo C (2014) Phosphoproteomic analysis of the non-seed vascular plant model Selaginellamoellendorffii. Proteome Sci 12:16. doi:10.1186/1477-5956-12-16
Curaba J, Talbot M, Li Z, Helliwell C (2013) Over-expression of microRNA171 affects phase transitions and floral meristem determinancy in barley. BMC Plant Biol 13:6. doi:10.1186/1471-2229-13-6
Devers EA, Teply J, Reinert A, Gaude N, Krajinski F (2013) An endogenous artificial microRNA system for unraveling the function of root endosymbioses related genes in Medicagotruncatula. BMC Plant Biol 13:82. doi:10.1186/1471-2229-13-82
Dugas DV, Bartel B (2004) MicroRNA regulation of gene expression in plants. Curr Opin Plant Biol 7(5):512–520. doi:10.1016/j.pbi.2004.07.011
Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, Carrington JC (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One 2(2):e219. doi:10.1371/journal.pone.0000219
FdeF Rosas-Cárdenas, Duran-Figueroa N, Vielle-Calzada JP, Cruz-Hernandez A, Marsch-Martinez N, de Folter S (2011) A simple and efficient method for isolating small RNAs from different plant species. Plant Methods 7:4. doi:10.1186/1746-4811-7-4
Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N (2012) miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucl Acids Res 40(1):37–52. doi:10.1093/nar/gkr688
Gandikota M, Birkenbihl RP, Höhmann S, Cardon GH, Saedler H, Huijser P (2007) The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J 49(4):683–693. doi:10.1111/j.1365-313X.2006.02983.x
Gao Z, Shi T, Luo X, Zhang Z, Zhuang W, Wang L (2012) High-throughput sequencing of small RNAs and analysis of differentially expressed microRNAs associated with pistil development in Japanese apricot. BMC Genom 13:371. doi:10.1186/1471-2164-13-371
Garcia D (2008) A miRacle in plant development: role of microRNAs in cell differentiation and patterning. Semin Cell Dev Biol 19:586–595. doi:10.1016/j.semcdb.2008.07.013
Ghatnekar L, Jaarola M, Bengtsson BO (2006) The introgression of a functional nuclear gene from Poa to Festucaovina. Proc R Soc B 273:395–399. doi:10.1098/rspb.2005.3355
Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A (2005) Rfam: annotating non-coding RNAs in complete genomes. Nucl Acids Res 33:D121–D124. doi:10.1093/nar/gki081
Griffiths-Jones S, Saini HK, Van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36:D154–D158. doi:10.1093/nar/gkm952
Gu M, Liu W, Meng Q, Zhang W, Chen A, Sun S, Xu G (2014) Identification of microRNAs in six solanaceous plants and their potential link with phosphate and mycorrhizal signaling. J Integr Plant Biol 56(12):1164–1178. doi:10.1111/jipb.12233
Guo L, Chen F (2014) A challenge for miRNA: multiple isomiRs and miRNAomics. Gene 544:1–7. doi:10.1016/j.gene.2014.04.039
Guo HS, Xie Q, Fei JF, Chua NH (2005) MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17(5):1376–1386. doi:10.1105/tpc.105.030841
Hausler RE, Baur B, Scharte J, Theichmann T, Eicks M, Fischer KL, Flugge UI, Schubert S, Weber A, Fischer K (2000) Plastidic metabolite transporters and their physiological functions in the inducible crassulacean acid metabolism plant Mesembryanthemum crystallinum. Plant J 24(3):285–296. doi:10.1046/j.1365-313x.2000.00876.x
Hong Y, Jackson S (2015) Floral induction and flower formation—the role and potential applications of miRNAs. Plant Biotechnol J 13(3):282–292. doi:10.1111/pbi.12340
Huijser P, Schmid M (2011) The control of developmental phase transitions in plants. Development 138(19):4117–4129. doi:10.1242/dev.063511
Jin Q, Xue Z, Dong C, Wang Y, Chu L, Xu Y (2015) Identification and characterization of MicroRNAs from Tree Peony (Paeoniaostii) and their response to copper stress. PLoS One 10(2):e0117584. doi:10.1371/journal.pone.0117584
Kalve S, De Vos D, Beemster GT (2014) Leaf development: a cellular perspective. Front Plant Sci 5:362. doi:10.3389/fpls.2014.00362.eCollection2014
Karlova R, van Haarst JC, Maliepaard C, van de Geest H, Bovy AG, Lammers M, Angenent GC, de Maagd RA (2013) Identification of microRNA targets in tomato fruit development using high-throughput sequencing and degradome analysis. J Exp Bot 64(7):1863–1878. doi:10.1093/jxb/ert049
Kim HJ, Baek KH, Lee BW, Choi D, Hoor CG (2011) In silico identification and characterization of microRNAs and their putative target genes in Solanaceae plants. Genome 54(2):91–98. doi:10.1139/G10-104
Knox JP (1995) The extracellular matrix in higher plants. 4. Developmentally regulated proteoglycans and glycoproteins of the plant cell surface. FASEB J 9(11):1004–1012
Leake JR (1994) The biology of myco-heterotrophic (‘saprophytic’) plants. New Phytol 127:171–216. doi:10.1111/j.1469-8137.1994.tb04272.x
Lewis LA, Mccourt RM (2004) Green algae and the origin of land plants. Am J Bot 91(10):1535–1556. doi:10.3732/ajb.91.10.1535
Li X, Hou Y, Zhang L, Zhang W, Quan C, Cui Y, Bian S (2014) Computational identification of conserved microRNAs and their targets from expression sequence tags of blueberry (Vaccinium corybosum). Plant Signal Behav 9(9):e29462. doi:10.4161/psb.29462
Liang H, Zen K, Zhang J, Zhang CY, Chen X (2013) New roles for microRNAs in cross-species communication. RNA Biol 10(3):367–370. doi:10.4161/rna.23663
Liu PP, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC (2007) Repression of auxin response factor10 by microRNA160 is critical for seed germination and post-germination stages. Plant J 52(1):133–146. doi:10.1111/j.1365-313X.2007.03218.x
Liu Q, Yao X, Pi L, Wang H, Cui X, Huang H (2009a) The ARGONAUTE 10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. Plant J 58(1):27–40. doi:10.1111/j.1365-313X.2008.03757.x
Liu Q, Zhang YC, Wang CY, Luo YC, Huang QJ, Chen SY, Zhou H, Qu LH, Chen YQ (2009b) Expression analysis of phytohormoneregulated microRNAs in rice, implying their regulation roles in plant hormone signaling. FEBS Lett 583(4):723–728. doi:10.1016/j.febslet.2009.01.020
Liu Q, Wang H, Hu H, Zhang H (2015) Genome-wide identification and evolutionary analysis of positively selected miRNA genes in domesticated rice. Mol Genet Genomics 290(2):593–602. doi:10.1007/s00438-014-0943-0
Lu SF, Sun YH, Shi R, Clark C, Li LG, Chiang VL (2005a) Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 17(8):2186–2203. doi:10.1105/tpc.105.033456
Lu C, Tej SS, Luo SJ, Haudenschild CD, Meyers BC, Green PJ (2005b) Elucidation of the small RNA component of the transcriptome. Science 309(5740):1567–1569. doi:10.1126/science.1114112
Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, Griffiths-Jones S, Jacobsen SE, Mallory AC, Martiessen RA, Poethig RS, Qi Y, Vaucheret H, Voinnet O, Watanabe Y, Weigel D, Zhu JK (2008) Criteria for annotation of plant MicroRNAs. Plant Cell 20(12):3186–3190. doi:10.1105/tpc.108.064311
Miyashima S, Koi S, Hashimoto T, Nakajima K (2011) Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138:2303–2313. doi:10.1242/dev.060491
Molnar A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC (2007) miRNAs control gene expression in the single-cell alga Chlamydomonasreinhardtii. Nature 447:1126–1129. doi:10.1038/nature05903
Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC (2008) Specificity of argonaute7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133(1):128–141. doi:10.1016/j.cell.2008.02.033
Morin RD, Aksay G, Dolgosheina E, Ebhardt HA, Magrini V, Mardis ER, Sahinalp SC, Unrau PJ (2008) Comparative analysis of the small RNA transcriptomes of Pinuscontorta and Oryza sativa. Genome Res 18(4):571–584. doi:10.1101/gr.6897308
Okoye IS, Coomes SM, Pelly VS, Czieso S, Papayannopoulos V, Tolmachova T, Seabra MC, Wilson MS (2014) MicroRNA-containing T-regulatory-cell-derived exosomes suppress pathogenic T helper 1. Cells Immun 41(1):89–103. doi:10.1016/j.immuni.2014.05.019
Ori N, Cohen AR, Etzioni A, Brand A, Yanai O, Shleizer S, Menda N, Amsellem Z, Efroni I, Pekker I, Alvarez JP, Blum E, Zamir D, Eshed Y (2007) Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet 39(6):787–791. doi:10.1038/ng2036
Palmer CM, Hindt MN, Schmidt H, Clemens S, Guerinot ML (2013) MYB10 and MYB72 are required for growth under iron-limiting conditions. PLoSGenet 9(11):e1003953. doi:10.1371/journal.pgen.1003953
Pei H, Ma N, Chen J, Zheng Y, Tian J, Li J, Zhang S, Fei Z, Gao J (2013) Integrative analysis of miRNA and mRNA profiles in response to ethylene in rose petals during flower opening. PLoS ONE 8(5):e64290. doi:10.1371/journal.pone.0064290
Peiter E, Maathuis FJM, Mills LN, Knight H, Pelloux J, Hetherington AM, Sanders D (2005) The vacuolar Ca2+-activated channel TPC1 regulates germination and stomatal movement. Nature 434:404–408. doi:10.1038/nature03381
Peláez P, Trejo MS, Iñiguez LP, Estrada-Navarrete G, Covarrubias AA, Reyes JL, Sanchez F (2012) Identification and characterization of microRNAs in Phaseolus vulgaris by high-throughput sequencing. BMC Genom 13:83. doi:10.1186/1471-2164-13-83
Qiu D, Pan X, Wilson IW, Li F, Liu M, Teng W, Zhang B (2009) High throughput sequencing technology reveals that the taxoid elicitor methyl jasmonate regulates microRNA expression in Chinese yew (Taxuschinensis). Gene 436(1–2):37–44. doi:10.1016/j.gene.2009.01.006
Rajagopalan R, Vaucheret H, Trejo J, Bartel D (2006) A diverse and evolutionarily fluid set of micro-RNAs in Arabidopsis thaliana. Genes Dev 20(24):3407–3425. doi:10.1101/gad.1476406
Rajwanshi R, Chakraborty S, Jayanandi K, Deb B, Lightfoot DA (2014) Orthologous plant microRNAs: microregulators with great potential for improving stress tolerance in plants. Theor Appl Genet 127(12):2525–2543. doi:10.1007/s00122-014-2391-y
Reyes JL, Chua NH (2007) ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J 49(4):592–606. doi:10.1111/j.1365-313X.2006.02980.x
Rubio-Somoza I, Weigel D (2011) MicroRNA networks and developmental plasticity in plants. Trends Plant Sci 16(5):258–264. doi:10.1016/j.tplants.2011.03.001
Shalom L, Shlizerman L, Zur N, Doron-Faigenboim A, Blumwald E, Sadka A (2015) Molecular characterization of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) gene family from Citrus and the effect of fruit load on their expression. Front Plant Sci 6:389. doi:10.3389/fpls.2015.00389
Singh R, Jamieson A, Cresswell P (2008) GILT is a critical host factor for Listeria monocytogenes infection. Nature 455(7217):1244–1247. doi:10.1038/nature07344
Soltis DE, Albert VA, Leebens-Mack J, Palmer JD, Wing RA, dePamphilis CW, Ma H, Carlson JE, Altman N, Kim S, Wall PK, Zuccolo A, Soltis PS (2008) The Amborella genome: an evolutionary reference for plant biology. Genome Biol 9(3):402. doi:10.1186/gb-2008-9-3-402
Song C, Wang C, Zhang C, Korir NK, Yu H, Ma Z, Fang J (2010) Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata). BMC Genom 11:431. doi:10.1186/1471-2164-11-431
Stocks MB, Moxon S, Mapleson D, Woolfenden HC, Mohorianu I, Folkes L, Schwach F, Dalmay T, Moulton V (2012) The UEA sRNA workbench: a suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics 28(15):2059–2061. doi:10.1093/bioinformatics/bts311
Subramanian S, Fu Y, Sunkar R, Barbazuk WB, Zhu JK, Yu O (2008) Novel and nodulation-regulated microRNAs in soybean roots. BMC Genom 9:160. doi:10.1186/1471-2164-9-160
Sunkar R, Jagadeeswaran G (2008) In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol 8:37. doi:10.1186/1471-2229-8-37
Szittya G, Moxon S, Santos DM, Jing R, Fevereiro MP, Moulton V, Dalmay T (2008) High-throughput sequencing of Medicagotruncatula short RNAs identifies eight new miRNA families. BMC Genom 9:593. doi:10.1186/1471-2164-9-593
Tatematsu K, Toyokura K, Miyashima S, Nakajima K, Okada K (2015) A molecular mechanism that confines the activity pattern of miR165 in Arabidopsis leaf primordia. Plant J 82(4):596–608. doi:10.1111/tpj.12834
Taylor RS, Tarver JE, Hiscock SJ, Donoghue PC (2014) Evolutionary history of plant microRNAs. Trends Plant Sci 19:175–182. doi:10.1016/j.tplants.2013.11.008
Teotia S, Tang G (2015) To Bloom or Not to Bloom: role of MicroRNAs in Plant Flowering. Mol Plant 8(3):359–377. doi:10.1016/j.molp.2014.12.0187
The Gene Ontology Consortium (2008) The gene ontology project in 2008. Nucleic Acids Res 36:D440–D444. doi:10.1093/nar/gkm883
Tian DQ, Pan XY, Yu YM, Wang WY, Zhang F, Ge YY, Shen XL, Shen FQ, Liu XJ (2013) De novo characterization of the Anthurium transcriptome and analysis of its digital gene expression under cold stress. BMC Genom 14:827. doi:10.1186/1471-2164-14-82
Trindade I, Capitão C, Dalmay T, Fevereiro MP, Santos DM (2010) miR398 and miR408 are up-regulated in response to water deficit in Medicagotruncatula. Planta 231:705–716. doi:10.1007/s00425-009-1078-0
Umen JG (2014) Green algae and the origins of multicellularity in the plant kingdom. Cold Spring Harb Perspect Biol 6:a016170. doi:10.1101/cshperspect.a016170
Vasudevan S, Tong Y, Steitz JA (2007) Switching from repression to activation: microRNAs can up-regulate translation. Science 318(5858):1931–1934. doi:10.1126/science.1149460
Vaucheret H, Vazquez F, Crete P, Bartel DP (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18(10):1187–1197. doi:10.1101/gad.1201404
Vidal EA, Araus V, Lu C, Parry G, Green PJ, Goruzzi GM, Gutierrez RA (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl AcadSci USA 107(9):4477–4482. doi:10.1073/pnas.0909571107
Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136(4):669–687. doi:10.1016/j.cell.2009.01.046
Wang JW, Wang LJ, Mao YB, Cai WJ, Xue HW, Chen XY (2005) Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17(8):2204–2216. doi:10.1105/tpc.105.033076
Wang CY, Chen YQ, Liu Q (2011a) Sculpting the meristem: The roles of miRNAs in plant stem cells. Biochem Biophys Res Commun 409(3):363–366. doi:10.1016/j.bbrc.2011.04.123
Wang Y, Itaya A, Zhong X, Wu Y, Zhang J, van der Knaap E, Olmstead R, Qi Y, Ding B (2011b) Function and evolution of a MicroRNA that regulates a Ca2+-ATPase and triggers the formation of phased small interfering RNAs in tomato reproductive growth. Plant Cell 23(9):3185–3203. doi:10.1105/tpc.111.088013
Wu MF, Tian Q, Reed JW (2006) Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133(21):4211–4218. doi:10.1242/dev.02602
Xia R, Zhu H, An YQ, Beers EP, Liu Z (2012) Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol 13(6):R47. doi:10.1186/gb-2012-13-6-r47
Xia R, Meyers BC, Liu Z, Beers EP, Ye S, Liu Z (2013) MicroRNA superfamilies descended from miR390 and their roles in secondary small interfering RNA Biogenesis in Eudicots. Plant Cell 25(5):1555–1572. doi:10.1105/tpc.113.110957
Yang JH, Han SJ, Yoon EK, Lee WS (2006) Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res 34(6):1892–1899. doi:10.1093/nar/gkl118
Yang ZB, Eticha D, Albacete A, Rao IM, Roitsch T, Horst WJ (2012) Physiological and molecular analysis of the interaction between aluminium toxicity and drought stress in common bean (Phaseolus vulgaris). J Exp Bot 63(8):3109–3125. doi:10.1093/jxb/ers038
Yant L, Mathieu J, Dinh TT, Ott F, Lanz C, Wollmann H, Chen X, Schmid M (2010) Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22(7):2156–2170. doi:10.1105/tpc.110.075606
Yin Z, Li C, Han X, Shen F (2008) Identification of conserved microRNAs and their target genes in tomato (Lycopersiconesculentum). Gene 414(1–2):60–66. doi:10.1016/j.gene.2008.02.007
Zhang B, Pan X, Cobb GP, Anderson TA (2006a) Plant microRNA: a small regulatory molecule with big impact. Developmental Biology 289(1):3–6. doi:10.1016/j.ydbio.2005.10.036
Zhang BH, Pan XP, Cox SB, Cobb GP, Anderson TA (2006b) Evidence that miRNAs are different from other RNAs. Cell Mol Life Sci 63:246–254. doi:10.1007/s00018-005-5467-7
Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA (2006c) Conservation and divergence of plant microRNA genes. Plant J 46:243–259. doi:10.1111/j.1365-313X.2006.02697.x
Zhang Z, Yu J, Li D, Zhang Z, Liu F, Zhou X, Wang T, Ling T, Su Z (2010) PMRD: plant microRNA database. Nucl Acids Res 38:D806–D813. doi:10.1093/nar/gkp818
Zhang H, Zhao X, Li J, Cai H, Deng XW, Li L (2014) MicroRNA408 is critical for the HY5-SPL7 gene network that mediates the coordinated response to light and copper. Plant Cell 26(12):4933–4953. doi:10.1105/tpc.114.127340
Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y (2007) A complex system of small RNAs in the unicellular green alga Chlamydomonasreinhardtii. Genes Dev 21(10):1190–1203
Zhong B, Sun L, Penny D (2015) The origin of land plants: a phylogenomic perspective. Evol Bioinform 11:137–141. doi:10.4137/EBO.S29089
Zhu QH, Helliwell CA (2011) Regulation of flowering time and floral patterning by miR172. J Exp Bot 62(2):487–495. doi:10.1093/jxb/erq295
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31(13):3406–3415. doi:10.1093/nar/gkg595
Acknowledgments
This work was supported by a Grant from the Russian Science Foundation (14-24-00175), and was done with the use of the experimental climate control facility.
Author’s contribution
Conceived and designed the experiments: N.V.R., E.B.P., K.G.S. Performed the experiments: A.M.M., E.Z.K. Analyzed the data: A.V.B., A.V.S., O.A.S. Contributed reagents, materials, and analysis tools: N.V.R, E.B.P, A.V.B. Provided plant material: E.Z.K. Wrote the paper: A.V.S. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM_1 Supplementary Table S1
Summary of high-throughput sequencing of M. hypopitys small RNA library (PDF 164 kb)
ESM_2 Supplementary Table S2
Known mhy-miRNAs and their targets in M. hypopitys (XLSX 49 kb)
ESM_3 Supplementary Table S3
M. hypopitys miRNA isoforms (PDF 273 kb)
ESM_4 Supplementary Table S4
Identified precursors of known and candidate novel mhy-miRNAs, and novel mhy-miRNA targets (XLSX 24 kb)
ESM_5 Supplementary Figure S1
Secondary pre-miRNA structures of known conserved (1A - 1D) and candidate novel (1E - 1U) mhy-miRNAs. 1A shows pre-mhy-miR116a; 1B - pre-mhy-miR167a; 1C - pre-mhy-miR393ab; 1D - pre-mhy-miR1094c; 1E - pre-mhy-miR0001; 1F - pre-mhy-miR0007; 1G - pre-mhy-miR0008; 1H - pre-mhy-miR0009; 1I - pre-mhy-miR0013; 1 J - pre-mhy-miR0017; 1 K - pre-mhy-miR0026; 1L - pre-mhy-miR0036; 1 M - pre-mhy-miR0037; 1 N - pre-mhy-miR0039; 1O - pre-mhy-miR0044; 1P - pre-mhy-miR0045; 1Q - pre-mhy-miR0046; 1R - pre-mhy-miR0048; 1S - pre-mhy-miR00053; 1T - pre-mhy-miR0054; 1U - pre-mhy-miR0060 (PDF 1165 kb)
ESM_6 Supplementary Table S5
Selected target genes of M. hypopitys miRNAs compared to known plant miRNAs (PDF 309 kb)
Rights and permissions
About this article
Cite this article
Shchennikova, A.V., Beletsky, A.V., Shulga, O.A. et al. Deep-sequence profiling of miRNAs and their target prediction in Monotropa hypopitys . Plant Mol Biol 91, 441–458 (2016). https://doi.org/10.1007/s11103-016-0478-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-016-0478-3