Abstract
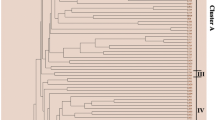
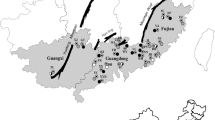
Some more than a century old Chinese fir genotypes with high biomass that are now rare in China were historically called “the king of Chinese fir”. The genetic diversity and relationships of those ancient Chinese fir genotypes were investigated using morphological analysis and sequence-related amplified polymorphism markers. The morphological analysis indicated that the tree volume parameter had the maximum variable coefficient value (70.2 %). In total, 18 selected primers generated 154 bands, and 150 bands were polymorphic (97.4 %). Higher values of genetic diversity parameters (h = 0.3593, I = 0.5283) were maintained at the species level, indicating that the ancient Chinese fir in China has retained a relatively high level of genetic diversity. AMOVA indicated that 73.16 % of the variation resided within provenances. The UPGMA dendrogram and genetic structure analysis identified 3 major clusters and grouped the genotypes in agreement with their geographic origins. A Mantel test revealed a significant correlation between genetic distances and geographic distances among the genotypes (r = 0.4275, p < 0.01). Overall, we recommend a combination of conservation measures including a germplasm repository and the implementation of in situ conservation.



Similar content being viewed by others
References
Aneja B, Yadav NR, Chawla V, Yadav RC (2012) Sequence-related amplified polymorphism (SRAP) molecular marker system and its applications in crop improvement. Mol Breed 30:1635–1648
Bi J, Blanco J, Seely B, Kimmins J, Ding Y, Welham C (2007) Yield decline in Chinese-fir plantations: a simulation investigation with implications for model complexity. Can J For Res 37:1615–1630
Bian L, Shi J, Zheng R, Chen J, Wu HX (2014) Genetic parameters and genotype–environment interactions of Chinese fir (Cunninghamia lanceolata) in Fujian Province. Can J For Res 44:582–592
Chen L, Chen F, He S, Ma L (2014) High genetic diversity and small genetic variation among populations of Magnolia wufengensis (magnoliaceae), revealed by ISSR and SRAP markers. Electron J Biotechnol 17:268–274
Deng WZ (2011) Effects of reforestation methods on forest soil properties in mid-subtropical mountainous area in the south of China. J Subtrop Res Environ 6:18–23
Duan AG, Zhang XQ, Zhang JG, Zhong JD (2014) Growth and genetic evaluation of 21-year-old Chinese Fir clonal plantation. For Res 27:672–676 (in Chinese)
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Fu YB (2015) Understanding crop genetic diversity under modern plant breeding. Theor Appl Genet 128:2131–2142
Guo J, Yang Y, Chen G, Xie J, Gao R, Qian W (2010) Effects of clear-cutting and slash burning on soil respiration in Chinese fir and evergreen broadleaved forests in mid-subtropical China. Plant Soil 333:249–261
He G, Xu Y, Qi M, Shen F, Zhang J, Luo W (2011) Genetic variation and individual plant selection of main economic traits of progenies from the second-generation seed orchards of Chinese Fir. For Res 24:123–126 (in Chinese)
Huang Z, He Z, Wan X, Hu Z, Fan S, Yang Y (2013) Harvest residue management effects on tree growth and ecosystem carbon in a Chinese fir plantation in subtropical China. Plant Soil 364:303–314
Jump AS, Marchant R, Peñuelas J (2009) Environmental change and the option value of genetic diversity. Trends Plant Sci 14:51–58
Laidò G, Mangini G, Taranto F, Gadaleta A, Blanco A, Cattivelli L, Marone D, Mastrangelo AM, Papa R, De VP (2013) Genetic diversity and population structure of tetraploid wheats (Triticum turgidum L.) estimated by SSR, DArT and pedigree data. PLoS One 8:1591–1594
Li M, Shi J, Li F, Gan S (2007) Molecular characterization of elite genotypes within a second-generation Chinese Fir (Cunninghamia lanceolata) breeding population using RAPD markers. Scientia Silvae Sinicae 43:50–55 (in Chinese)
Li M, Zhao Z, Miao X (2014) Genetic diversity and relationships of apricot cultivars in North China revealed by ISSR and SRAP markers. Sci Hortic 173:20–28
Liedloff A (1999) MantelV2.0, nonparametric test calculator. Queensland University of Technology, Australia
Ma X, Wu P, Huang M, Chen X, Zou X, Chen N (2014) The kings of Chinese Fir. Chinese Forestry Publishing House, pp 1–15 (in Chinese)
Ming L, Zhong Z, Xingjun M, Jingjing Z (2013) Genetic diversity and population structure of Siberian apricot (Prunus sibirica L.) in China. Int J Mol Sci 15:377–400
Nei M (1978) The theory of genetic distance and evolution of human races. Jpn J Hum Genet 23:341–369
Ouyang L, Chen J, Zheng R, Xu Y, Lin Y, Huang J, Ye D, Fang Y, Shi J (2014) Genetic diversity among the germplasm collections of the Chinese fir in 1st breeding population upon SSR markers. J Nanjing For Univ 1:21–26 (in Chinese)
Pauls SU, Nowak C, Balint M, Pfenninger M (2013) The impact of global climate change on genetic diversity within populations and species. Mol Ecol 22:925–946
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 7:574–578
Robarts DW, Wolfe AD (2014) Sequence-related amplified polymorphism (SRAP) markers: a potential resource for studies in plant molecular biology. Appl Plant Sci 2:15–18
Rohlf FJ (2000) NTSYS-PC, numerical taxonomy system for the PC Exeter Software, Version 2.1. Applied Biostatistics Inc Setauket, USA
Shen AH, Li HB, Wang K, Ding HM, Zhang X, Fan L, Jiang B (2011) Sequence characterized amplified region (SCAR) markers-based rapid molecular typing and identification of Cunninghamia lanceolata. Afr J Biotechnol 10:19066–19074
Sheng WT, Wang L, Zhang HY (1981) A preliminary study on the climatic regions of Chinese Fir growth areas. Scientia Silvae Sinicae 17:50–57 (in Chinese)
Tong CF, Shi JS (2004) Constructing genetic linkage maps in Chinese Fir using F1 progeny. Yi Chuan Xue Bao 31:1149–1156 (in Chinese)
Wan LC, Wang F, Guo X, Lu S, Qiu Z, Zhao Y, Zhang H, Lin J (2012) Identification and characterization of small non-coding RNAs from Chinese fir by high throughput sequencing. BMC Plant Biol 12:1–15
Wang Q, Wang S, Feng Z, Deng S, Gao H (2004) An overview on studies of soil organic matter in Chinese fir plantation. Yingyong Shengtai Xuebao 15:1947–1952 (in Chinese)
Wang D, Wang B, Dai W, Li P, Hu W, Guo H (2009) The variation characteristics of soil organic carbon and its influence factor in different developing stages of Chinese Fir plantations. For Res 22:667–671 (in Chinese)
Wu PF, Tigabu M, Ma XQ, Oden PC, He YL, Yu XT, He ZY (2011) Variations in biomass, nutrient contents and nutrient use efficiency among Chinese fir provenances. Silvae Genet 60:95–105
Wu Z, Zhou C, Zhou X, Zheng L, Lai A, Lu X (2014) Study on the ecological response of Cunninghamia lanceolata plantation to selective cutting intensity in Mountain South China. In: Lee G (ed) Environment and sustainability, vol 154. WIT Press, Southampton, p 267
Xie Q, Liu ZH, Wang SH, Li ZQ (2015) Genetic diversity and phylogenetic relationships among five endemic Pinus taxa (Pinaceae) of China as revealed by srap markers. Biochem Syst Ecol 62:115–120
Xu Y, Chen J, Zhao Y, Wang Y, Wang X, Liu W, Shi J, Zheng R, Ouyang L, Zhang Z (2014) Variation of EST-SSR molecular markers among provenances of Chinese fir. J Nanjing For Univ 38:1–8 (in Chinese)
Yang YS, Guo JF, Chen GS, He ZM, Xie JS (2003) Effect of slash burning on nutrient removal and soil fertility in Chinese fir and evergreen broadleaved forests of mid-subtropical China. Pedosphere 13:87–96
Yang YS, Guo JF, Chen GS, Xie JS, Gao R, Li Z, Jin Z (2005) Carbon and nitrogen pools in Chinese fir and evergreen broadleaved forests and changes associated with felling and burning in mid-subtropical China. For Ecol Manag 216:216–226
Yang YL, Xiang Qing MA, Zhang MQ (2009) Molecular polymorphic analysis for different geographic provenances of Chinese Fir. J Trop Subtrop Botany 17:183–189 (in Chinese)
Yeh FC, Yang RC, Boyle TB, Ye Z, Mao JX (1997) POPGENE, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Centre, University of Alberta, Edmonton
Yu X (1997) Cultivation of Chinese Fir. Fujian Science and Technology Press, Fuzhou, pp p1–p30
Zhang P, Yang L (2014) Review of study on the impact of China’s forest certification. For Econ 8:103–108 (in Chinese)
Zhanjun W, Jinhui C, Weidong L, Zhanshou L, Pengkai W, Yanjuan Z, Renhua Z, Jisen S (2013) Transcriptome characteristics and six alternative expressed genes positively correlated with the phase transition of annual cambial activities in Chinese Fir (Cunninghamia lanceolata (Lamb.) Hook). PLoS ONE 8:e71562
Zheng R (2005) Research advances in genetic breeding of Chinese Fir. World For Res 18:63–65 (in Chinese)
Zheng HQ, Duan HJ, Hu DH, Wei R, Li Y (2015a) Sequence-related amplified polymorphism primer screening on Chinese fir (Cunninghamia lanceolata (Lamb.) Hook). J For Res 26:101–106
Zheng HQ, Duan HJ, Hu DH, Li Y, Hao YB (2015b) Genotypic variation of Cunninghamia lanceolata revealed by phenotypic traits and SRAP markers. Dendrobiology 74:85–94
Zheng HQ, Hu DH, Wang RH, Wei RP, Yan S (2015c) Assessing 62 Chinese Fir (Cunninghamia lanceolata) breeding parents in a 12-year grafted clone test. Forests 6:3799–3808
Zhiwen LI, Baosheng LI, Sun L, Wang F (2015) Paleontological evidence of the changes of the north boundary of southern mid-subtropical zone in eastern China during holocene. Trop Geogr 35:179–185 (in Chinese)
Acknowledgments
We acknowledge the financial support of the National Natural Science Foundation of China (U1405211, 31500541).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, M., Chen, X., Huang, M. et al. Genetic diversity and relationships of ancient Chinese fir (Cunninghamia lanceolata) genotypes revealed by sequence-related amplified polymorphism markers. Genet Resour Crop Evol 64, 1087–1099 (2017). https://doi.org/10.1007/s10722-016-0428-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-016-0428-6