Abstract
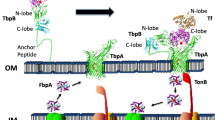
Lactoferrin (LF) is a member of the transferrin family that is abundantly expressed and secreted by glandular epithelial cells. The biological functions of LF involve in iron homeostasis regulation of the body and antibacterial activity. Previous studies demonstrated that it had a high cationic N-terminal domain that could interact with glycosaminoglycans, lipopolysaccharides and the bacterial virulence protein. Two anti-microbial peptides, lactoferricin (LFcin) and lactoferrampin (LFampin), were also isolated and identified in N-terminal of LF. Although the antibacterial mechanism was carefully studied, little was known about the molecular evolution of LF. In this study, we estimated the nonsynonymous-to-synonymous substitution ratios (\( \omega = {{d_{N} } \mathord{\left/ {\vphantom {{d_{N} } {d_{S} }}} \right. \kern-\nulldelimiterspace} {d_{S} }} \)) per site using maximum likelihood method to analyze the LF evolution. The results of ω > 1 and five identified positive selection sites of amino acid suggested that the evolution of LF gene was characterized by positive selection. Further study found that the positive selection sites were either located in the LF-bacteria binding region or the peptides of LFcin and LFampin, indicating that the selection pressure was related to LF-bacteria interaction. The identification of these sites may contribute to the mechanism of bacteria-LF interaction.




Similar content being viewed by others
References
Aagaard JE, Phillips P (2004) Accuracy and power of the likelihood ratio test for comparing evolutionary rates among genes. J Mol Evol 60:426–433
Baker EN, Baker HM (2009) A structural framework for understanding the multifunctional character of lactoferrin. Biochimie 91:3–10
Baker HM, Baker CJ, Smith CA et al (2000) Metal substitution in transferrins: specific binding of cerium (IV) revealed by the crystal structure of cerium-substituted human lactoferrin. J Biol Inorg Chem 5:692–698
Bellamy W, Takase M, Yamauchi K et al (1992) Identification of the bactericidal domain of lactoferrin. Biochim Biophys Acta 1121:130–136
Bolscher GMJ, Adao R, Nazmi K et al (2009) Bactericidal activity of LFchimera is stronger and less sensitive to ionic strength than its constituent lactoferricin and lactoferrampin peptides. Biochimie 91:123–132
Editorial (2009) Advances in lactoferrin research. Biochimie 91:1–2
Ford MJ (2001) Molecular evolution of transferrin: evidence for positive selection in Salmonids. Mol Biol Evol 18:639–647
Gifford JL, Hunter HN, Vogel HJ (2005) Lactoferricin: a lactoferrin-derived peptide with anti-microbial, antiviral, antitumor and immunological properties. Cell Mol Life Sci 62:2588–2598
Haney EF, Nazmi K, Lau F et al (2009) Novel lactoferrampin antimicrobial peptides derived from human lactoferrin. Biochimie 91:141–154
Haridas M, Anderson BF, Baker EN (1995) Structure of human diferric lactoferrin refined at 2.2 Å resolution. Acta Cryst D51:629–646
Havard J, Robert EWH (2009) Antimicrobial properties of lactoferrin. Biochimie 91:19–29
Karthikeyan S, Paramasivam M, Yadav S et al (1999) Structure of buffalo lactoferrin at 2.5 Å resolution using crystals grown at 303 K shows different orientations of the N and C lobes. Acta Cryst D55:1805–1813
Khan JA, Kumar P, Paramasivam M et al (2001) Camel lactoferrin-a transferrincum-lactoferrin: crystal structure of camel apolactoferrin at 2.6 Å resolution and structural basis of its dual role. J Mol Biol 309:751–761
Lambert AL, Perri H, Meehan TJ (2005a) Evolution of duplications in the transferrin family of proteins. Comp Biochem Physiol, Part B 140:11–25
Lambert AL, Perri H, Halbrooks JP et al (2005b) Evolution of the transferrin family: Conservation of residues associated with iron and anion binding. Comp Biochem Physiol, Part B 142:129–141
Moore SA, Anderson BF, Groom CR et al (1997) Three dimensional structure of diferric bovine lactoferrin at 2.8 Å resolution. J Mol Biol 274:222–236
Nielsen R, Yang Z (1998) Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148:929–936
Pettersen EF, Goddard TD, Huang CC et al (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Retzer MD, Yu R, Schryvers AB (1999) Identication of sequences in human transferring that bind to the bacterial receptor protein, transferrin-binding protein B. Mol Microbiol 32:111–121
Sharma AK, Paramasivam M, Srinivasan A et al (1998) Three-dimensional structure of mare diferric lactoferrin at 2.6 Å resolution. J Mol Biol 289:303–317
Susana AGC, Sigifredo AG, Quintín RC (2009) Lactoferrin: structure, function and applications. Int J Antimicrob Agents 33:301.e1–301.e8
Tamura K, Dudley J, Nei M et al (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Trinchella F, Parisi E, Scudiero R (2009) Evolutionary analysis of the transferrin gene in Antarctic Notothenioidei: a history of adaptive evolution and functional divergence. Marine Genomics 1:95–101
Valenti P, Antonini G (2005) Lactoferrin: an important host defence against microbial and viral attack. Cell Mol Life Sci 62:2576–2587
Van der Kraan MIA, Groenink J, Nazmi K et al (2004) Lactoferrampin: a novel antimicrobial peptide in the N1-domain of bovine lactoferrin. Peptides 25:177–183
Wong WSW, Yang Z, Goldman N et al (2004) Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics 168:1041–1051
Yang Z (2006) Computational molecular evolution. Oxford University Press, New York
Yang Z (2007) PAML4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Yang L, Gui JF (2004) Positive selection on multiple antique allelic lineages of transferrin in the polyploid carassius auratus. Mol Biol Evol 21:1264–1277
Yang Z, Nielsen R, Goldman N et al (2000) Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–449
Yang Z, Wong WSW, Nielsen R (2005) Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol 22:1107–1118
Acknowledgments
We thank Professor Rasmus Nielsen of Biology at the University of Copenhagen who gave Jiang many suggestions concerning this research. This work was supported by National Natural Science Foundation of China grant 30300253 and the earmarked fund for Modern Agro-industry Technology Research System to Professor X.P. Jiang.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liang, G.M., Jiang, X.P. Positive selection drives lactoferrin evolution in mammals. Genetica 138, 757–762 (2010). https://doi.org/10.1007/s10709-010-9456-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-010-9456-x