Abstract
Context
Previous studies have raised concerns about the analysis and meta-analysis of crossover experiments and we were aware of several families of experiments that used crossover designs and meta-analysis.
Objective
To identify families of experiments that used meta-analysis, to investigate their methods for effect size construction and aggregation, and to assess the reproducibility and validity of their results.
Method
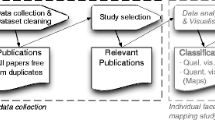
We performed a systematic review (SR) of papers reporting families of experiments in high quality software engineering journals, that attempted to apply meta-analysis. We attempted to reproduce the reported meta-analysis results using the descriptive statistics and also investigated the validity of the meta-analysis process.
Results
Out of 13 identified primary studies, we reproduced only five. Seven studies could not be reproduced. One study which was correctly analyzed could not be reproduced due to rounding errors. When we were unable to reproduce results, we provide revised meta-analysis results. To support reproducibility of analyses presented in our paper, it is complemented by the reproducer R package.
Conclusions
Meta-analysis is not well understood by software engineering researchers. To support novice researchers, we present recommendations for reporting and meta-analyzing families of experiments and a detailed example of how to analyze a family of 4-group crossover experiments.
Similar content being viewed by others
1 Introduction
Vegas et al. (2016) reported that crossover designs are a popular design for software engineering experiments. In their review they identified 82 papers of which 33 (i.e., 40.2%) were crossover designs. Furthermore, those 82 papers reported 124 experiments of which 68 (i.e., 54.8%) used crossover designs. However, they reported that “crossover designs are often not properly designed and/or analysed, limiting the validity of the results”. They also warned against the use of meta-analysis in the context of crossover style experiments.
As a results of that study, two of us undertook a detailed study of parametric effect sizes from AB/BA crossover studies (see Madeyski and Kitchenham 2018a, band Kitchenham et al. 2018). We identified the need to consider two mean difference effect sizes and reported the small sample effect size variances and their normal approximations.
As we were undertaking this systematic review,Footnote 1 we found that Santos et al. (2018) had already performed a mapping study of families of experiments. They reported that although the most favoured means of aggregating results was Narrative synthesis (used by 18 papers), Aggregated Data meta-analysis (by which they mean aggregation of experiment effect sizes) was used by 15 studies.
Using Vegas et al. (2016), Madeyski and Kitchenham (2018b) and Santos et al. (2018) as a starting point, we decided to investigate the validity and reproducibility of effect size meta-analysis for families of experiments (Madeyski and Kitchenham 2017). Our goals are to;
Identify the effect sizes used and how they were calculated and aggregated.
Use the descriptive statistics reported in the study, attempt to reproduce the reported results.Footnote 2
In the event that we were unable to reproduce the results, to investigate the underlying reason for lack of reproduciblity.
We concentrated on families of experiments as our form of primary studies. We did this (rather than looking at papers that report a meta-analysis after performing a systematic review) because papers reporting a family of experiments are likely to have published sufficient details about the individual studies and their meta-analysis process for us to attempt to validate and reproduce their effect size calculations and meta-analysis. In addition, Santos’s mapping study confirmed the popularity of families of experiments, and emphasized that more families needed to aggregate their results. These two factors indicate the importance of adopting valid meta-analysis processes in the context of families of experiments. Nonetheless, our reproducibility analysis method, based on aggregating descriptive statistics, is the same as would be used to meta-analyse data from experiments found by a systematic review. Thus, the results from this study are likely to be of value for any meta-analysis of software engineering data.
We concentrated on high quality journals not only because such papers usually present reasonably complete descriptions of their results and methods, but also because they attract papers from experienced researchers, which are reviewed by other experienced researchers. Thus, readers of papers in such journals expect the published results to be correct. Invalid results in such papers are therefore likely to have a more serious impact than mistakes in papers published in less prestigious journals or conferences. For example, practitioners may base decisions on invalid outcomes, and novice researchers may adopt incorrect methods.
We present our research questions in Section 2 and our systematic review methods in Section 3. A summary of the primary studies included in our review, a discussion of the validity of the meta-analysis methods used in each study and our reproducibility assessment are in Sections 4, 5 and 6, respectively. We discuss the results of our study in Section 7 and present the contributions of this paper and our conclusions in Section 8.
We also include an Appendix that reports details of our statistical analysis and analysis results not needed to support our main arguments. The Appendix also discusses reproducibility aspects of our study.
2 Research Questions
The research questions (RQs) relating to our systematic review are:
- RQ1:
Which studies that undertook families of experiment have also undertaken effect size meta-analysis?
- RQ2:
What are the characteristics of these studies in terms of methods used for experimental design and analysis?
- RQ3:
What meta-analysis methods were used and were they valid?
- RQ4:
If the meta-analysis methods were valid can results be successfully reproduced?
RQ1, RQ2, and the reporting aspects of RQ3 could be addressed directly from information reported in each primary study. To address the validity aspect of RQ3 and RQ4, we reviewed the meta-analysis processes described by each study and then attempted to reproduce first the effect sizes and then the meta-analysis in each primary study. Finally, we compared our results with the reported results. We assumed that it would be possible to conduct a meta-analysis based on the descriptive data and the effect size chosen by the primary study authors, since this is the normal method of performing meta-analysis.
3 Systematic Review Methods
We performed our systematic review (SR) according to the guidelines proposed by Kitchenham et al. (2015). The processes we adopted are specified in the following sections.
3.1 Protocol Development
Our protocol defines the procedures we intended to use for the systematic review including the search process, the primary study selection process, the data extraction process and the data analysis process. It also identified the main tasks of all the co-authors. The protocol was initially drafted by the first author and reviewed by all the authors. After trialling the specified processes, the final version of the protocol was agreed by all the authors and registered as report W08/2017/P-045 at Wroclaw University of Science and Technology. The following sections are based on the processes defined in the protocol. Any divergences report our actual processes, as opposed to the planned processes described in the protocol. The major deviation from the protocol and the results reported in this paper is that originally we had assumed it would be appropriate to concentrate on reproducibility, but as our investigation progressed we realized that we needed to consider the reasons for lack of reproducibility, that is, consider in more detail the validity of the meta-analysis process. Furthermore, validity is the key issue, because it is not useful to reproduce an invalid result.
3.2 Search Strategy
In order to address our research questions, we needed to identify papers that reported the use of meta-analysis to aggregate individual studies, reported the results of the individual studies in detail, and were published in high quality journals.
To achieve our search process strategy, we decided to limit our search for families of experiments to the following five journals:
IEEE Transactions on Software Engineering (TSE).
Empirical Software Engineering (EMSE).
Journal of Systems and Software (JSS).
Information and Software Technology (IST).
ACM Transactions on Software Engineering Methodology (TOSEM).
We restricted ourselves to these journals because they all publish papers on empirical software engineering, and all have relatively high impact factors (among SE journals). These are, therefore, highly respected journals, and we should expect the quality of papers they publish to be correspondingly high.
3.3 SR Inclusions and Exclusions
In this section we present our inclusion and exclusion criteria. Details of the search and selection process, the validation of the search and selection process, and the data extraction process can be found in the supplementary material (Kitchenham et al. 2019b).
Given our research questions, papers to be included in our SR were identified using the following inclusion criteria:
- 1.
The paper should report a family of three or more experiments. This is because it is the criteria adopted by Santos et al. (2018) and there is more opportunity to detect heterogeneity with three or more studies.
- 2.
The experiments reported in the paper should relate to human-centric experiments or quasi-experiments that compare SE methods or procedures rather than report observational (correlation) studies with no clear comparisons.Footnote 3
- 3.
The paper should have been published by one of the five journals identified by our search strategy, see Section 3.2.
- 4.
The paper should use some form of meta-analysis to aggregate results from the individual studies using standardized effect sizes, i.e., standardized mean difference or point-biserial correlation coefficient (rpb).Footnote 4 These effect sizes are commonly used in software engineering meta-analyses.
The following exclusion criteria were also defined:
- 1.
The paper was an editorial.Footnote 5
- 2.
The paper was published before 1999, when Basili et al. (1999) first discussed families of experiments.
3.4 Data Analysis
The results extracted from each primary study allowed us to answer questions RQ1, RQ2 and the methodology element of RQ3. To address the validity element of RQ3 and RQ4 for each primary study, we reviewed carefully the meta-analysis methods reported by the study authors and attempted to reproduce the effect size values and meta-analysis results using the reported descriptive data.
Many of the studies reported multiple metrics and hypotheses tests for each experiment. In all cases, we first attempted to reproduce the effect sizes reported by the authors and then the meta-analysis. We analyzed only the first outcome metric, because we assumed that if the individual effect sizes were reproduced and results of meta-analyzing the effect sizes was reproduced, it would confirm whether or not the meta-analysis was reproducible without checking the results for every metric. Our assumption (that in our case it is enough to analyze the first outcome metric) was based on the fact that none of the primary studies reported using different methods to calculate effect sizes or performing meta-analysis for different outcome metrics. In addition, outcome tables for descriptive statistics and effect sizes were similar for all outcome metrics. There is only one situation where there might be a difference between outcomes for different metrics. This would happen if the authors did not maintain the direction as well as the magnitude of the effect size. Then, if one metric had effect sizes with different directions and one did not, we would agree with the authors in the case where all directions were the same and disagree when the directions were not the same. This happened in the case of Study 9 (see Section 6.11).
For each primary study, we compared the effect sizes for each experiment and the overall meta-analysis mean effect size with the results of our calculations. However, we needed some method of deciding whether effect sizes or meta-analysis results had been reproduced, since we did not expect to obtain exactly the same effect size values since our values were obtained from summary statistics whereas study authors might have derived their effect sizes from calculations on the raw data. We chose to use a difference of 0.05 between our calculated effect size meta-analysis mean and the equivalent reported statistics as a criterion for deciding whether there was a reproducibility problem. Our basis for choosing 0.05 was that:
- 1.
A relative value would unfairly penalize small effect sizes, for example if a study reported an effect size of 0.01 and we reported an effect size of 0.02, we would have relative difference of 50% for a difference that could be the result of rounding applied to reported mean values.
- 2.
Most studies reported descriptive data on metrics, in the range 0 to 1, to two decimal places, so we thought an absolute value of 0.05 might be sufficiently large to allow for differences due to rounding effects caused because our reproducibility statistics were derived from the reported means and variances.
- 3.
Most studies did not state explicitly whether or not they applied the small sample size adjustment to their standardized effect sizes. For example, a medium effect size of 0.5 and a sample size of 23 (the median experiment size), the effect of applying the small sample adjustment is to reduce the standardized effect size to 0.48.
4 An Overview of the Primary Studies (RQ1 and RQ2)
In this section, we address RQ1 and RQ2 and present an overview of the primary studies included in our systematic review.
4.1 Studies Reporting Meta-analysis of Families of Experiments (RQ1)
The 13 primary studies we included in our SR are shown in Table 1 ordered by inverse publication date.Footnote 6 The table reports the number of experiments in each family and the number of participants in each experiment. We report on the studies in this order throughout this section.
Table 2 provides an overview of the goals of each of the studies and the specific techniques they investigated. The technique in boldface (e.g., PBR in study S13) is the treatment technique and the other technique (e.g., CBR) is the control technique. Later in this paper, effect sizes are reported relative to the treatment technique, so positive values indicate that the treatment technique outperforms the control technique and negative values indicate that the control technique outperforms the treatment technique. There are some trends observable in Table 2:
Six studies investigated the impact of different UML documentation options (see rows where the techniques are labelled DO to signify Documentation Options).
Four studies investigated procedures in the context of maintainability.
Four studies investigated requirements issues, three compared specification languages and one investigated proposals for verifying non-functional requirements.
4.2 Experimental Methods Used by the Primary Studies (RQ2)
Table 3 presents some information about individual experiments discussed in each primary study. During data extraction, it became clear that many of our 13 primary studies, included experiments with crossover designs. Vegas et al. (2016) warned that the terminology used to describe crossover designs was not used consistently, and we found exactly the same problem with our primary studies (Kitchenham et al. 2019a). Therefore, we used the description of the experimental design provided by the authors to derive our own classification. Understanding the specific experimental design is important in the context of meta-analysis, because the variance of the standardized effect size is different for different designs, see Morris and DeShon (2002) and Madeyski and Kitchenham(2018a, b). In all cases the description was sufficient for us to identify the individual experimental designs. Like Vegas et al., we found that the primary study authors did not adopt our terminology, nor did they use the same terminology as other primary study authors who adopted the same design.
The primary studies used only four basic experimental designs, which we discuss in the Appendix A.1. To understand the notation used in the rest of the paper, it is important to note that all crossover style designs have two different types of standardized mean difference effect size (see Morris and DeShon 2002 and Madeyski and Kitchenham 2018b):
- 1.
An effect size that measures the personal improvement (of an individual or team) performing a task using one method compared with performing the same taskFootnote 7 using another method. We refer to this as the repeated measures standardized effect size, δRM, with an estimate dRM.
- 2.
An effect size that is equivalent to the standardized mean effect size obtained from a an independent groups design (also known as a between participants design). We refer to this independent groups effect size as δIG, with an estimate dIG.
For balanced crossovers (where each sequence group has the same number of participants), effect sizes are calculated as follows (Morris and DeShon 2002; Madeyski and Kitchenham 2018b):
where \(\bar {x}_{A}\) is the mean value of the treatment technique observations and \(\bar {x}_{B}\) is the mean value of control technique, se is the within participants standard deviation.
where sIG is equivalent to the pooled within groups standard deviation of an independent groups study.
In addition, there is a relationship between the two standard deviations (Madeyski and Kitchenham 2018b):
where r is the Pearson correlation between the repeated measures. Thus, the effect sizes are also related:
For small sample size, Hedges and Olkin (1985) recommend applying a correction to dRM and dIG. We refer to the small sample size corrected effect sizes as gRM and gIG respectively. We prefer not to give these terms generic labels, such as Hedges’ g, because as Cumming (2012) points out (see page 295) meta-analysis terminology is inconsistent. In terms of names given to standardized effect sizes, dIG is referred to as d by Borenstein et al. (2009) and as g by Hedges and Olkin (1985), gIG is referred to as g by Borenstein et al. (2009) and d by Hedges and Olkin (1985). In our primary studies, most papers used the terms Hedge’s g and one used Cohen’s d but the papers did not specify whether or not they used the small sample size adjustment. Only Study 13, explicitly defined Hedges’ g to be what we refer to as dIG and used the term d to be what we refer to as gRM.
In Table 3, we also report whether the data was analyzed using parametric (P) or non-parametric methods (NP) tests for the individual experiments. Four of the studies used non-parametric tests or parametric tests depending on the outcome of tests for normality. Study 13 and Study 14 performed both non-parametric and parametric tests, but only reported the results of the parametric tests since the outcomes of both tests were consistent. It is important to note that many of the crossover studies did not analyze their data correctly, by using independent groups tests rather than repeated measures tests. We annotated three studies as partly valid because they used tests that catered for repeated measures, but may have been delivered slightly biased results if time period effects or material effects were significant (see Appendix A.1.3).
5 The Validity of Meta-analysis Procedures Used by the Primary Studies (RQ3)
In this section, we discuss the methods used by the primary study authors. In Table 4, we summarize issues related to meta-analysis including the effect size names used by the authors, our assessment of the effect size the authors aggregated, which meta-analysis tools were used and whether heterogeneity was investigated. We discuss these results in this section. However, the main focus of this section is to assess the validity of the meta-analysis procedures used in each primary study. This validity assessment was made from reading the report of the meta-analysis processes and the meta-analysis results reported in each primary study. It was intended to identify incorrect or incomplete reporting of meta-analysis process and any obvious violations of meta-analysis principles. In Section 5.1, we explain the recommended methods for analyzing standardized mean difference effect sizes, then in Section 5.2, we discuss the methods used by the primary study authors and highlight any potential validity problems with their meta-analysis method.
5.1 Standard Procedures for Meta-analysis
The usual method for aggregating standardized mean effect sizes such as Hedges’ g is to construct a weighted average using the inverse of the effect size variance: (see, for example, Hedges and Olkin 1985; Lipsey and Wilson 2001; Borenstein et al.2009):
where ESi is the calculated effect size of the i-th experiment, k is the number of experiments, \(\overline {ES}\) is the mean effect size, and wi is an appropriate weight. It is also customary to use the inverse of the effect size variance as the weight, i.e., wi = 1/(var(ES)i), where the formula for (var(ES)i) depends both on the study design (Morris and DeShon 2002; Madeyski and Kitchenham 2018b) and the specific effect size. However, Hedges and Olkin (1985) make it clear that the use of the variance is based on large sample theory. In practice using the estimate of ESi in the equation for its variance, when sample sizes are small, leads to a biased weights and a biased estimate of \(\overline {ES}\). They point out that a weight based on the number of observationsFootnote 8 would lead to a pooled estimate that was unbiased but less precise. Such weights are close to optimal when the population mean is close to zero and the number of observations are large.
Equation (5) assumes a fixed effects meta-analysis but a random effects analysis is also usually based on the effect size variance. Also, in the case of a fixed effect analysis, the variance of \(\overline {ES}\) is obtained from the equation:
Equation (5) is also used for aggregating the unstandardized effect size (UES). Although in this case, var(UES)i is the square of the standard error of the mean difference.
There are two main meta-analysis models: a fixed effects model and a random effects model. Equations (5) and (6) are appropriate for a fixed effects model, when we assume that data from individual experiments arise from the same population (i.e., the data from each experiment arise from the same population).
A random effects model assumes that data from individual experiments arise from different populations each of which has its own population mean and variance. A random effects analysis estimates the excess variance due to the different populations by comparing the variance between experiment means with the within experiment variance. In practice, random effects analysis replaces var(ES)i with a larger revised variance that includes both the within experiment variance and the between experiment variance. In the case of a family of experiments, we would expect a priori that the experiments were closely controlled replications and a fixed effect size would be appropriate. However, a random effects analysis will give the same results as a fixed effects analysis in the event that the effect sizes are homogeneous, so we would recommend defaulting to a random effects method. Such approach would address the common issue, also mentioned by Santos et al. (2018), of using fixed effect models when, due to the heterogeneity of effects, random effects models would be preferred.
5.2 Meta-analysis Methods Used by the Primary Studies
None of the primary studies aggregated the unstandardized effect size. However, twelve studies reported effect sizes they referred to either as Hedges’ g or a related standardized effect size (Cohen’s d, γ and d). Apart from Study 13, none of the papers that used crossover-style experiments mentioned the possibility of two different effect sizes, so we assume that they all attempted to aggregate the effect size equivalent to an independent group study (i.e., dIG or gIG).
Study 1 and Study 4 both reported calculating Hedges’ g, but their description did not mention applying the small sample size adjustment, so we assume they reported what we refer to as dIG. They also reported converting to a correlation based effect size (usually referred to as the point bi-serial correlation, rpb Rosenthal 1991). This can easily be calculated from the standardized effect size using the following formula (see Borenstein et al. 2009; Lipsey and Wilson 2001):
where a = 4 for a balanced experiment. After constructing rpb, it is necessary to apply Fisher’s normalising transformation Fisher (1921). The resulting transformed variable for experiment i is referred to as zi, and the set of zi −values can be aggregated using the following equation (which is equivalent to (5)):
The only mistake Study 1 and Study 4 made in the description of their meta-analysis was that the authors reported using a weight wi = 1/(N − 3), where wi is the weight for the i th experiment. In fact, the variance of rpb, after applying the Fisher normalizing transformation, is vi = 1/(N − 3) and the weight is wi = 1/vi = (N − 3), which ensures that the largest studies are given most weight in the aggregation process (Lipsey and Wilson 2001). In addition, the authors of Study 4 reported using a t-test for independent groups, so they may have used the number of observations rather than the sample size to calculate weights (and the overall variance).
In principle, transformation to rpb is a valid analysis method, since it avoids the probable bias in calculating the variance of the dIG for small sample sizes. For this reason, we used it as the basis of our reproducability analysis, and we report the method in detail in Appendix A.2.
An important implication of using the normalizing transformation of rpb is that the variance of rpb is var(ri) = 1/(ni − 3) and using (6):
This means that if researchers mistakenly believe the variance is based on the number of observations rather than the number of participants, they will assume that the variance of each rpb is 1/(2ni − 3) after transformation, and will substantially underestimate the variance of the average effect size \(\overline {r_{pb}}\).
Four studies (i.e., Study 2, Study 5, Study 9 and Study 10) reported an effect size that they referred to as Hedges’ g. They also reported an aggregation method that, like Study 1 and Study 4, used (8), and they also made the same mistake with their description of the weight. However, they did not explicitly confirm that they transformed their effect size to a correlation, so we cannot be sure whether these studies aggregated the standardized effect sizes directly but mistakenly assumed that the variance of each effect size was 1/(ni − 3), or omitted to mention that they used the rpb transformation. Of these four studies, only Study 2 used an analysis that considered repeated values, so the other studies might have used a variance based on 1/(2ni − 3).
Study 3, Study 7 and Study 11 all made a mistake with their basic meta-analysis. They all used an AB/BA crossover design (although Study 3 also used an independent groups design for one of its 5 experiments). In each crossover study they estimated a standardized effect size for each time period separately. So for each AB/BA experiment they calculated two different estimates of dIG, one for time period 1 and the other for time period 2. It is incorrect to aggregate such effect sizes because the same participants contributed to each estimate of dIG, and, hence, the two effect sizes from the same experiment were not independent. This violates one of the basic assumptions of meta-analysis that each effect size comes from an independent experiment. The effect of this error is to increase the degrees of freedom attributed to tests of significance associated with the average effect size.
Study 6 reported using Cohen’s d and aggregating their values using a weighted mean and the META 5.3 tool. They referenced Hedges and Olkin (1985), which did not report methods for meta-analysing crossover designs, so we assume that the authors aggregated dIG but do not know how they calculated their weights.
Study 8 reported and aggregated rpb but used a different method to that used by Study 1 and Study 4. We describe the method they used in the Appendix A.3. From the viewpoint of validity a critical issue is that they derived rpb from the one-sided p −value of their statistical tests. For each experiment in the family and for each metric, they used either the Mann-Whitney-Wilcoxon (MMW) test or the t −test depending on the outcome of a normality test. However, Study 8 used statistical tests appropriate for independent groups studies, although the family used 4-group crossover experiments, so the resulting p −values are likely to be invalid. However, the study authors were attempting to use a meta-analysis process that would allow them to aggregate their parametric and non-parametric results. The authors reported the heterogeneity of their experiments, but as pointed out in Appendix A.3, the heterogeneity was probably over-estimated.
Study 13 reported a standardized effect size based on team improvement, which we refer to as gRM. The authors also reported dIG for each experiment, which they referred to as Hedges’ g, but they did not aggregate it. They estimated the variance of dRM but do not cite the origin of the formula they used. They used Hedges’ Q statistic (see (19)) to test for heterogeneity. The test failed to reject the null hypothesis (i.e., their p −value was greater than 0.05), and they reported what appears to be the unweighted mean of the effect sizes.
Study 14 referred to their effect size as γ for 4 separate hypotheses. However, the hypothesis we believe to be most relevant to investigating the difference between the techniques was based on the difference between the personal improvement observed among participants in one treatment group and the personal improvement among participants in the other group. This is a difference of differences analysis for which it is correct to use the independent groups t −test. However, γ cannot be easily equated to either dRM or dIG. For purposes of analysis, the difference data can be analysed as an independent groups study, but for purposes of interpretation, the mean difference measures the average individual improvement after the effect of skill differences are removed. They report both the weighted and unweighted overall mean. As explained in Appendix A.1.1, the weight was based on the inverse of the variance of γ and was calculated using the formula for the moderate sample-size approximation of the variance of gIG. They also tested for heterogeneity using the Q statistic proposed by by Hedges and Olkin (1985) which depends on the effect size variance.
Both Study 13 and Study 14 also aggregated one-sided p −values, as described in Appendix A.4, in order to test the null hypothesis of no significant difference between techniques.
The majority of primary study authors used the Meta-Analysis v2 BioStat (2006) for aggregation, although Meta-Analysis v2 does not support aggregation results from crossover design studies.
As mentioned by Santos et al. (2018), although many researchers used non-parametric methods for at least some of their individual experiments (see Table 3), they subsequently used parametric effect sizes. This is somewhat inconsistent but not necessarily invalid. It would certainly be inappropriate for studies that used both parametric and non-parametric methods to aggregate non-parametric effect sizes and parametric effect sizes in the same meta-analysis, so some consistent effect size metric is necessary.
The advantage of using the standardized mean difference is that the central limit theorem confirms that mean differences are normal irrespective of the underlying distribution of the data. The problem with standardized effect sizes is that the estimate of the variance of the data within each experiment, which is used to calculate the standardized effect size, may be biased for small sample sizes. However, the variance of the mean effect sizes for each experiment calculated as part of any random effects meta-analysis puts an upper limit on the variance of the overall mean effect size. In addition, currently, aggregating non-parametric effect sizes is not feasible. There are no well-defined guidelines identifying which non-parametric effect sizes to use, nor how they might be aggregated.
Only three of the primary studies considered heterogeneity. Study 8 and Study 13 reported non-significant heterogeneity. Study 14 reported significant heterogeneity and reported both a weighted and an unweighted mean. Only Study 2 explicitly mentioned using a fixed effects meta-analysis. Since the other studies made no mention of heterogeneity or using any specific meta-analysis model, we assume that the they also undertook fixed effects meta-analysis.
6 The Reproducibility and Validity of the Primary Study Meta-analyses (RQ4)
This section reports our reproducibility assessment and incorporates it with the validity analysis reported in Section 5, since it makes little sense to investigate the reproducibility of invalid meta-analyses. In turn, our reproducibility assessment allowed us to investigate further the validity of the meta-analysis processes adopted in each paper, from the viewpoint of whether processes that were valid in principle, were also applied correctly, in practice. In Section 6.1, we describe the method we used for our reproducibility assessment. In Section 6.2, we report the overall results of the reproducibility assessment, and in the following sections, we discuss the reproducibility results for each study in the context of the validity assessment reported in Section 5.2.
6.1 Reproducibility Assessment Process
For reproducibility, as far as possible, we used the same method for each study. To construct the effect size, we used the following process:
- 1.
From the descriptive statistics reported in the study, we used (2) to calculate the standardized effect size appropriate for independent groups dIG. Our estimate of \(s^{2}_{IG}\) was usually based on the pooled within-technique variance. However, in the case of Study 3, Study 7 and Study 11, \(s^{2}_{IG}\) was based on the pooled within-cell variance, where a cell is defined as a set of observations that were obtained under exactly the same experimental conditions (see Appendix A.1.2).
- 2.
We applied the exact small sample size adjustment J (see (14)) to calculate the effect size gIG.
This is the standard starting point for any meta-analysis when raw data is not available. To aggregate the effect sizes:
- 1.
We transformed the gIG values to rpb and applied Fisher’s normalizing transformation Fisher (1921).
- 2.
We used the Rmetafor tool Viechtbauer (2010) to fit a random effects model using its default method which is the Restricted maximum-likelihood estimation (RMLE) method.
- 3.
We back-transformed our meta-analysis results to the standardized mean difference.
This approach is described in more detail in the Appendix A.2. It was the same as that undertaken by Abrahão et al. (2011), which has the advantage of being appropriate for all experimental designs used in our primary studies and does not rely on information such as the variances of standardized effect sizes which was not well-known to SE researchers.
The three main deviations from this method were:
- 1.
For Study 8, we reported our results in terms of the point bi-serial correlation (i.e., rpb) because Study 8 reported and aggregated rpb.
- 2.
For Study 13, descriptive statistics were not reported explicitly and we estimated the mean difference and standard deviations from the reported graphics. In addition, Study 13 explicitly reported the statistics we refer to as gRM and dIG, so we reported both effect sizes and, like the study authors, aggregated the gRM values.
- 3.
In Study 14, the authors reported the personal improvement results for each participant, which is equivalent to dRM. So, to report comparable effect sizes, we calculated the descriptive statistics from the reported descriptive difference data (i.e., the post-training results minus the pre-training results).
Assuming the descriptive data was reported correctly, our meta-analyses should provide more trustworthy results for studies that used an invalid meta-analysis process (in particular, Study 3, Study 7 and Study 11). However, as explained in Appendix A.1.2, if materials, or time period effects are significant our estimates of \(s^{2}_{IG}\) will be inflated which would lead to underestimates of dIG. Also if there were significant interactions between either time period or materials, and technique such effects would also inflate \(s^{2}_{IG}\).
We defined results to be reproducible if the difference between the individual experiment effect sizes and the overall effect size reported in the primary study and those we calculated from the descriptive statistics was less than 0.05, as discussed in Section 3.4. We also compared the probability levels for the overall effect sizes. We expected primary studies that did not appreciate the impact of repeated measures would report smaller p −values than us. As discussed in Section 3.4, we only analyzed one measure per primary study.
6.2 Reproducibility Assessment Results
Table 5 displays the calculated effect sizes and reported effect sizes for each experiment and each effect size reported in each study. The variable Type refers to the effect size reported in the row. None of the studies apart from Study 7, Study 11 and Study 13 mentioned the small sample adjustment factor, so we assume that the standardized mean difference effect size reported by the authors is dRM. Study 13 reported both dIG and gRM, but aggregated gRM and the one-sided p −value. Study 7 and Study 11 reported two values that they called Hedges’ g. The value in their main tables was the small sample size adjusted standardized mean difference effect size, but they aggregated the non-adjusted effect size. The final column labelled RR (i.e., Results Reproduced) reports the number of times the absolute difference between the reported and calculated effect sizes was less than than 0.05 for all relevant entries. The studies for which all standardized effect sizes were reproduced are highlighted. We were only able to reproduce all standardized effect sizes for Study 2, Study 5 and Study 6, although for Study 14, we also reproduced the authors’ aggregation of p −values.
Table 6 displays the calculated and reported overall mean values for the effect sizes plus (if available) the p −value of the mean, the upper and lower confidence interval bounds (UB and LB), QE which is the heterogeneity test statistic and QEp which the the p −value of the heterogeneity statistic. The column RR identifies whether the difference between the calculated overall mean and the reported overall mean was greater than 0.05 (the studies for which this is the case are highlighted). The mean of the standardized effect sizes was reproduced for seven studies: Study 2, Study 5, Study 6, Study 8, Study 10, Study 11, and Study 13. However, Study 8 and Study 11 must be discounted because of validity problems.
The reproducibility results are collated with the validity assessment for each study, and are discussed in the following sections. In each section, the validity problems identified in Section 5 are identified in the paragraphs labelled “Meta-Analysis Validity Issue”. Critical issues that invalidate the aggregation performed by the authors are identified. If the reproducibility failed or was otherwise deemed invalid, we include a “Cause of Problem” paragraph. Validity issues identified as a result of our reproducibility assessment are identified as meta-analysis process implementation errors in the “Cause of Problem” paragraph.
6.3 Study 1 Validity and Reproducibility
- Meta-Analysis Method Validity Issues::
-
None.
- Author’s Aggregation Method::
-
Weighted mean of dRM based on transforming to and from rpb.
- Our Aggregation Method::
-
Weighted mean of gIG based on transforming to and from rpb as described in Appendix A.2.
- Individual Effect Size Reproducibility::
-
Failed.
- Mean Effect Size Reproducibility::
-
Failed.
- Cause of Problem::
-
Meta-analysis process implementation error - Incorrect use of meta-analysis tool.
Comments: Although we could not detect any validity problems with Study 1, and we based our meta-analysis on rpb derived from gIG, we could not reproduce the effect sizes nor the meta-analysis results. The study reported substantially smaller effect sizes, both for individual experiments and overall, than the ones we calculated. We contacted Prof. Abrahão who was the first author of this paper. She very kindly provided us with the raw data used in Study 1. Using Prof. Abrahão’s raw data, we recalculated gIG for each study and aggregated the data after transforming to rpb and following the process described in the Appendix A.5. Prof. Abrahão agreed with our analysis of her raw data. She also confirmed that she was attempting to calculate the matched pairs effect size (i.e., gRM).
The low values she obtained were due to several different factors. The most significant issue was that she used the Meta-Analysis-V2 tool BioStat (2006) that does not support crossover designs, although it does support matched pairs studies. The tool attempts to calculate gIG not gRM.Footnote 9
6.4 Study 2 Validity and Reproducibility
- Meta-Analysis Method Validity Issue 1::
-
It is unclear whether the paper aggregated the standardized effect size dIG directly or used the transformation to rpb.
- Meta-Analysis Method Validity Issue 2::
-
The weights and variances may have been based on the number of observations rather than the number of participants.
- Author’s Aggregation Method::
-
Unclear. Either the weighted mean of dIG based on transforming to and from rpb or the weighted mean of dIG with weight = N-3.
- Our Aggregation Method::
-
As for Study 1.
- Individual Effect Size Reproducibility::
-
Succeeded.
- Mean Effect Size Reproducibility::
-
Succeeded.
Comments: According to our criteria, Study 2 was fully reproduced with respect to the individual effect sizes and the weighted mean of the effect sizes. However, there is difference with respect to the p −values for the overall mean that is consistent with using the number of observations rather than the number of participants when calculating the variance of the effect size.
6.5 Study 3 Validity and Reproducibility
- Meta-Analysis Method Validity Issue 1::
-
Critical validity issue - Incorrect meta-analysis of non-independent effect sizes.
- Meta-Analysis Method Validity Issue 2::
-
Unclear whether the authors aggregated dIG or rpb.
- Meta-Analysis Method Validity Issue 3::
-
The weights and variances may have been based on the number of observations rather than the number of participants for AB/BA crossover experiments.
- Author’s Aggregation Method::
-
Unclear. Either the weighted mean of dIG based on transforming to and from rpb or the weighted mean of dIG with weight = N-3.
- Our Aggregation Method::
-
As for Study 1.
- Individual Effect Size Reproducibility::
-
Failed (4), Succeeded (1).
- Mean Effect Size Reproducibility::
-
Failed.
- Cause of Problem::
-
Critical validity issue.
Comments: Study 3 used different experiment designs. Four experiments were AB/BA crossover experiments, the fifth experiment was an independent groups study. We were able to reproduce the effect size for the fifth experiment.
It is important to note that even though Study 3 used two different experimental designs, once comparable effect sizes are constructed, in this case gIG, results from all experiments can be aggregated. Thus, we provide corrected effect sizes and an overall meta-analysis, using the reported descriptive statistics to calculate gIG for each experiment, followed by aggregation of normalized rpb values.
6.6 Study 4 Validity and Reproducibility
- Meta-Analysis Method Validity Issues::
-
The study might have based weights and variances on the number of observations rather than the number of participants.
- Author’s Aggregation Method::
-
Weighted mean of dIG based on transforming to and from rpb.
- Our Aggregation Method::
-
As for Study 1.
- Individual Effect Size Reproducibility::
-
Failed.
- Mean Effect Size Reproducibility::
-
Failed.
- Cause of Problem::
-
Meta-analysis process implementation error - Incorrect use of meta-analysis tool
Comments: Like Study 1, Study 4 reported transforming its standardized effect size to rpb but could not be reproduced. Like Study 1, it reported significantly smaller effect sizes, both for individual experiments and overall, than the ones we calculated. Prof. Abrahão was a co-author of this paper, but she informed us that the raw data for Study 4 were no longer available. However, since the pattern of results was similar to Study 1 (i.e., the experiment effect sizes were smaller than the one we calculated), it is likely that the analysis suffered from the same problems.
6.7 Study 5 Validity and Reproducibility
- Meta-Analysis Method Validity Issue 1::
-
The study might have based weights and variances on the number of observations rather than the number of participants.
- Meta-Analysis Method Validity Issue 2::
-
Unclear whether the authors aggregated dIG or rpb.
- Author’s Aggregation Method::
-
Unclear. Either the Weighted mean of dIG based on transforming to and from rpb or the weighted mean of dIG with weight=N-3.
- Our Aggregation Method::
-
As for Study 1.
- Individual Effect Size Reproducibility::
-
Succeeded.
- Mean Effect Size Reproducibility::
-
Succeeded.
Comments: Despite uncertainty about which effect size was aggregated, Study 5 was successfully reproduced both at the individual experiment level and at the overall meta-analysis level. The largest discrepancy occurred for the first experiment results. This was due to a probable rounding error. The mean values of Ueffec for the first experiment (E-UL) in Table 7 of Fernández-Sáez et al. (2016) are 0.76 for Low LoD and 0.76 for High LoD, so we calculated the mean difference (and the effect size) to be zero. In fact, Study 5 reports a standardized effect size of − 0.046 (see Fernández-Sáez et al. 2016, Fig. 4).
Study 5 did not explicitly report the confidence intervals on mean standardized effect size, but visual inspection of their forest plot (Fernández-Sáez et al. 2016, Fig. 4) suggests an interval of approximately [− 0.25, 0.4] which is smaller than the interval we calculated [− 0.343,0.612]. So, Study 5 might have underestimated the standard error of the mean standardized effect size.
6.8 Study 6 Validity and Reproducibility
- Meta-Analysis Method Validity Issue::
-
The study might have based weights and variances on the number of observations rather than the number of participants.
- Aggregation Method::
-
Based on dIG but not specified in detail.
- Our Aggregation Method::
-
As for Study 1.
- Individual Effect Size Reproducibility::
-
Succeeded.
- Mean Effect Size Reproducibility::
-
Succeeded.
Study 6 was successfully reproduced both for individual effect sizes and for the overall mean effect sizes. All discrepancies appear to have occurred because we calculated the small sample size adjusted values. The non-adjusted values for the three experiments are Exp1 = 0.579, Exp2 = 0.3517 and Exp3 = 0.5793, which are very close to the reported values.
6.9 Study 7 Validity and Reproducibility
- Meta-Analysis Method Validity Issue::
-
Critical validity issue - Incorrect meta-analysis of non-independent effect sizes.
- Author’s Aggregation Method::
-
Weighted mean of dIG for each time period.
- Our Aggregation Method::
-
As for Study 1.
- Individual Effect Size Reproducibility::
-
Failed
- Mean Effect Size Reproducibility::
-
Failed.
- Cause of Problem::
-
Critical validity issue.
Comments: Like Study 3, Study 7 calculated standard effect sizes separately for each study. Since the meta-analysis aggregation was invalid, we report our estimates of the effect sizes for each experiment and their overall mean.
We note, however, that the first time period analysis the authors performed is a valid independent groups analysis (see Senn 2002, Section 3.1.2), so a meta-analysis, based on all participants provides valid estimate of dIG and its variance. Compared with an analysis of data from both time periods, the analysis is based on one set of materials rather than two and the estimate of dIG may be biased if the randomization to groups was not sufficient to balance out skill differences. However, it is not affected by any technique by time period or technique by order interactions.
6.10 Study 8 Validity and Reproducibility
- Meta-Analysis Method Validity Issue 1::
-
Wrongly used p −values from independent groups tests to calculate rpb
- Meta-Analysis Method Validity Issue 2::
-
Used the number of observations in their heterogeneity assessment instead of the number of participants.
- Author’s Aggregation Method::
-
Weighted mean of rpb based on the Hunter-Schmidt method (Hunter and Schmidt 1990).
- Our Aggregation Method::
-
Aggregation of rpb derived from gIG.
- Individual Effect Size Reproducibility::
-
Failed.
- Mean Effect Size Reproducibility::
-
Succeeded due to accidental correctness.
- Cause of Problem::
-
Meta-analysis process implementation error - Inconsistency between reported p −values and calculated effect sizes.
Comments: Study 8 was reproduced for three of the four effect sizes and the overall mean. The largest discrepancy was found for the first experiment.
We based our estimate of rpb on the gIG, whereas the authors used (33), so discrepancies might have been due to the different methods of calculating rpb. Table 7 summarises our attempt to reproduce the effect size calculations used by the authors from the initial p −values. The p −values reported by the authors are shown in the first row with their equivalent Z −values in row 2. The first issue is that the p −value for the first experiment is large while the other p −values are small which leads to both positive and negative Z −values. The published box plots all had medians for the control that were smaller than the medians for the technique treatment, so we would expect all the studies to have small p −values for tests (assuming the authors calculated the probability that the control group exhibited larger values than the treatment group). Thus, it appears that value for p(Exp1) is anomalous and could be a typographical error. Furthermore, applying their procedure to the p −values, we did not obtain values of rpb any closer to their reported values than the values we obtained starting from our estimates of gIG, whether we used the number of observations (see row 4, rpb(NO)) or the number of participants (see row 5, rpb(NP)) in Table 7.
Thus, although the overall mean rpb value we obtained is very close to the overall mean reported by the authors, the process used to derive the individual effect sizes could not be reproduced.
6.11 Study 9 Validity and Reproducibility
- Meta-Analysis Method Validity Issue 1::
-
Unclear whether the authors aggregated dIG or rpb
- Meta-Analysis Method Validity Issue 2::
-
The study might have based weights and variances on the number of observations rather than the number of participants.
- Author’s Aggregation Method::
-
Unclear. Either the weighted mean of dIG based on transforming to and from rpb or the weighted mean of dIG with weight = N-3.
- Our Aggregation Method::
-
As for Study 1.
- Individual Effect Size Reproducibility::
-
Failed.
- Mean Effect Size Reproducibility::
-
Failed.
- Cause of Problem::
-
Meta-analysis process implementation error - Authors ignored effect size direction.
Comments: Study 9 was not reproduced either in terms of individual effect sizes or in terms of the overall mean. Looking at the effect sizes, it is clear that the authors of Study 9 aggregated the absolute mean effect sizes for each experiment, and so overestimated the overall effect size.
This is the only case in which it is possible for the results of a meta-analysis process using one metric to differ, with respect to reproducibility, from the the results obtained using another metric. If all effect sizes of the other metric were in the same direction, using the absolute effect size would not cause a reproducibility problem. This is in fact the case for the other metric used in this study.
6.12 Study 10 Validity and Reproducibility
- Meta-Analysis Method Validity Issue 1::
-
Unclear whether the authors aggregated dIG or rpb
- Meta-Analysis Method Validity Issue 2::
-
The study might have based weights and variances on the number of observations rather than the number of participants.
- Author’s Aggregation Method::
-
Unclear. Either the weighted mean of dIG based on transforming to and from rpb or the weighted mean of dIG with weight = N-3.
- Our Aggregation Method::
-
As for Study 1.
- Individual Effect Size Reproducibility::
-
Not reported.
- Mean Effect Size Reproducibility::
-
Succeeded.
Comments: Study 10 did not report individual experiment effect sizes, nor any p −values for the meta-analysis, but, did report an overall effect size very close to our calculation.
6.13 Study 11 Validity and Reproducibility
- Meta-Analysis Method Validity Issue::
-
Critical validity issue - Incorrect meta-analysis of non-independent effect sizes.
- Author’s Aggregation Method::
-
Weighted mean of dIG for each time period.
- Our Aggregation Method::
-
As for Study 1.
- Individual Effect Size Reproducibility::
-
Not reported.
- Mean Effect Size Reproducibility::
-
Succeeded due to accidental correctness.
- Cause of Problem::
-
Critical validity issue.
Comments: Like Study 3 and Study 7, Study 11 calculated standard effect sizes separately for each study. In this case, however, we found an example of accidental correctness. The Study 11 mean effect size was reproduced because the analysis effects were extremely close for both time periods so constructing an average effect size for each experiment gave very similar results to treating the results of each time periods as separate experiments. What is noticeable is that the reported p −value was considerably lower than the one we calculated. This was because the authors believed they had six effect sizes in their meta-analysis rather than three.
Like Study 7, the first time period meta-analysis reported by Study 11 provides a valid estimate of dIG and its variance.
6.14 Study 13 Validity and Reproducibility
- Meta-Analysis Method Validity Issue::
-
None
- Author’s Aggregation Method::
-
Unweighted mean of gRM and sum of the natural logarithm of the one-sided p −values.
- Our Aggregation Method::
-
Weighted mean of gRM based on transformation to and from rpb and sum of the natural logarithm of the one-sided p-values.
- Individual Effect Size Reproducibility::
-
Failed due to extracting basic data from graphics.
- Mean Effect Size Reproducibility::
-
Succeeded.
Comments: Study 13 did not report the mean and standard deviation of the technique groups. Instead, the authors presented the descriptive statistics in graphical form. However, in contrast to the other studies, Study 13 reported both the dIG (which they referred to as Hedges’ g) and gRM (which they referred to as d) using a valid formula to estimate its standard deviation.
Since the value we used to reproduce the effect sizes were estimated from a diagram, we expected the difference between our results and the reported results to be slightly larger than our 0.05 level, in fact all the differences were less than 0.08
Study 13 aggregated both the one-sided p-values and the individual gRM effect sizes. The overall mean gRM was validated by our difference criterion. The reported aggregated probability, P, was close to the value we calculated,Footnote 10 and overall we conclude that Study 13 has been successfully reproduced.
6.15 Study 14 Validity and Reproducibility
- Meta-Analysis Method Validity Issue::
-
None
- Author’s Aggregation Method::
-
Weighted and unweighted mean of gRM and sum of the natural logarithm of the one-sided p −values.
- Our Aggregation Method::
-
Weighted mean of gRM based on transformation to and from rpb and sum of the natural logarithm of the one-sided p-values.
- Individual Effect Size Reproducibility::
-
For gIG failed due to rounding errors, for p succeeded.
- Mean Effect Size Reproducibility::
-
Failed due to rounding errors.
Comments: Study 14 used an interesting design that avoids some of the problems associated with replicated measures by analyzing the differences in differences (see Appendix A.1.4). Study 14 actually performed four statistical tests for each of four different variables, including comparing the pretest results for each group, comparing the posttest results for each group, comparing the post-test with the pretest values for each group, as well comparing the mean difference of the difference between pretest and posttest results for each group (which they call the performance improvement). However, for the purpose of comparing the two treatments, the relative performance improvement is the most appropriate measure to test:
where xAi2 is the posttest value of metric x for participant i in Group A and xAi1 is the pretest value of metric x for subject i. xBi2 and xBi1 are equivalent values for participants in group B. nA and nB are the number of participants in each group. Like an independent groups analysis, the variance of the difference values is the pooled within group variance (see (12)).
We were able to reproduce only one of the standardized mean effect sizes for individual experiments. In addition, we could not reproduce the overall mean effect size. All the data is reported to two significant digits, and it appears that because the raw data values are quite small, this has led to potentially large rounding errorsFootnote 11 However, we obtained t-test p −values that were similar to the reported values, and our aggregated p −values were also close.
7 Discussion
This section discusses issues arising from our systematic review and validity and reproducibility studies.
7.1 Summary of Results
We found 13 primary studies that conformed with our inclusion criteria in the sources we searched. All primary studies reported their experimental designs in sufficient detail for us to classify their individual experiments into four distinct design types: 4-group AB/BA crossover design,duplicated AB/BA crossover design, independent groups design, and a pretest posttest control design.
All 13 primary studies also provided sufficient information for us to reproduce their meta-analysis results, but, in most cases, only for effects sizes comparable to independent groups designs (i.e., dIG and gIG). Of the crossover designs, only Study 13 reported the improvement effect sizes (gRM). The other crossover design studies did not provide the summary information needed to calculate the personal improvement effect size.
We identified four primary studies that exhibited validity problems sufficient to call into question the reported meta-analysis results, and another six studies where we were unsure about the validity of the meta-analysis. In those six cases, we expected the effect sizes to be slightly biased and effect size variances to be underestimated, see Appendix A.5 for a more detailed explanation.
Of the 12 studies that reported individual experiment effect sizes, we were able to fully reproduce five primary studies. In addition, we also reproduced six of the 12 reported overall effect sizes. In the case of Study 10, which did not report individual experiment effect sizes, we were able to reproduce its overall effect size.
7.2 Experimental Designs Used by Primary Studies
Six studies used the 4-group duplicated AB/BA crossover design and four studies used the AB/BA crossover design. Study 3 used two different designs, with 4 experiments using a 4-group duplicated AB/BA crossover and one experiment using an independent groups design. The two remaining studies used an independent groups design and a pretest posttest control design. Thus, 12 of the 13 primary studies used repeated measures methods.
Only one family used an independent groups design for all its experiments, although outcomes of this design are the most straightforward to analyse and meta-analyse. However, using more complex designs makes the analysis of individual experiments and their subsequent meta-analysis more difficult. Only 4 of those 12 repeated measures studies used analysis methods appropriate for repeated measures data. Using analysis methods appropriate for independent groups studies has knock-on effects for any subsequent meta-analysis that can lead to invalid effect sizes or invalid effect size variances.
The main reason for using repeated measures designs is to be able to account for the individual skill differences among participants. However, the crossover design is not the only way to do this. In particular, the pretest posttest control group experimental design (see Appendix A.1.4) has some desirable properties. It allows the effect of individual differences are catered for by the analysis, but avoids the problem of technique by period interaction which is a potential risk when using a crossover design. For example, there were many studies evaluating the perspective-based code reading (PBR) methods (see Ciolkowski 2009), some of which used the undefined current method as a control while others used the checklist-based reading (CBR) method as a control. Using a pretest posttest control group, the current method would be used to establish a pretest baseline and then groups could be randomly assigned to training in CBR or PBR and the posttest differences used to assess whether PBR or CBR most enhanced defect detection.
7.3 Meta-analysis Reporting
Primary study authors did not always describe their meta-analysis processes fully and consistently. Few studies reported any information related to the standard error of the average effect size or its confidence intervals. The p −values for the overall effect sizes were reported nine times. In only three cases were the reported and calculated p −values of the same order of magnitude. Two papers reported confidence interval bounds, but these were Study 7 and Study 11 and we disagreed with their aggregation process.Footnote 12
We also noticed some more general reporting issues:
Studies often reported a name such as Hedges’ g for their standardised mean effect sizes, but did not usually specify how this was calculated. For reproducibility it is important to know both the formula for the standard deviation used to standardise the mean difference and whether or not the small sample size adjustment factor was applied.
Many studies used metrics that corresponded to the fraction of correct responses and which they reported on a [0, 1] scale. This can lead to rounding errors when reproducing results, if descriptive statistics are only reported to two decimal places. It is preferable to represent such numbers as a percentage rather than a fractions. Reporting percentages to two decimal places is appropriate both for means and standard deviations.
Authors using a repeated measures design sometimes failed to report the number of participants in each sequence group. However, this is important for meta-analysis purposes if the individual experiments are unbalanced in any way.
We collate our observations and formulate guidelines about reporting and conduct of meta-analysis in Appendix A.6.
7.4 Meta-analysis Tools
11 of the 13 studies mentioned using a meta-analysis tool. Of those 11 studies, seven exhibited reproducibility problems. It is difficult for researchers to assess whether they have used tools correctly unless there is some way of validating the tool outcomes. This study has shown that attempting to reproduce the results from descriptive data is a useful means of checking the output from tools. Comparing the results of analyzing the raw data as opposed to the descriptive statistics (as reported in Appendix A.5) shows that results based on descriptive statistics may be biased, but they should still provide results of the same order of magnitude, providing a sanity check on the tool outputs.
7.5 Meta-analysis Methods
In this section we discuss the implications of our study on the use of meta-analysis methods to aggregate data from families of experiments.
7.5.1 Testing for Heterogeneity
Only three primary studies (Studies 8, 13 and 14) reported the results of testing for heterogeneity among experiments in a family. It might be expected that a family of experiments was by definition homogeneous. However, some studies such as Study 1 and Study 3 reported families that had considerable differences between the individual experiments (see the supplementary material (Kitchenham et al. 2019b)). It is certainly worth checking for heterogeneity in such cases. In the case of Study 1, our meta-analysis found a heterogeneity value of 4.01 which had an associated p −value of 0.45 suggesting that heterogeneity was limited and the fixed effect analysis undertaken by the authors was appropriate. In the case of Study 3, the heterogeneity value was 8.46 with p = 0.0761. Since heterogeneity tests are not very powerful (see Higgins and Thompson 2002), we suggest that a value less than 0.1 should be accepted as an indication that a random effects analysis might be preferable to a fixed effects analysis.
7.5.2 Meta-analysis Choices
One of the major problems with meta-analysis is that there are many different effect sizes and methods that can be used to aggregate results. The meta-analysis methods used in the primary studies were not always clearly reported, but most studies reported standardized mean effect sizes for individual effect sizes and for the overall mean effect size. Study 8 reported the point bi-serial correlation coefficient. In addition, Study 13 and 14 used the method of combining p-values, which is now known to have severe limitations, see Appendix A.4.
Many text books recommend aggregating standardised mean difference effect sizes, see for example, Borenstein et al. (2009) or Lipsey and Wilson (2001), but it depends on obtaining the correct effect size variance.Footnote 13 This is fairly straightforward if the individual experiments have medium to large sample sizes, but is more complicated if experiments have very small sample size (Hedges and Olkin 1985), and also depends on the specific experimental design, as can be seen in Madeyski and Kitchenham (2018b) and Morris and DeShon (2002).
It would seem to be easier to convert to rpb for aggregation, as we did in our reproducibility assessment. This procedure avoids the need to obtain estimates of the standardized effect size variance. However, it must be recognised that the problem with the standardised effect size and its variance is that, for small sample sizes, the estimate of the variance which is used to calculate the standardised effect size is likely to be inaccurate. Converting to rpb does not overcome this problem since the point bi-serial correlation is itself calculated as the ratio of two variance estimates.
In practice, as proposed by Santos et al. (2018), an option for homogeneous families (i.e., families that use the same material and the same output measures) would be to analyze the data from the family as one large experiment, using what they call an Independent Participant Data (IDP) stratified method. This analyzes the data from all the individual experiments together as a single data set, and uses the individual experiment identifier as a blocking factor. This would lead to an estimate of overall mean difference and the residual variance based on all the participants. An estimate of the effect size of the family and its standard error would then be more likely to be reliable.
It is also possible that using non-parametric effect sizes would avoid some of the problems inherent in using parametric effect sizes. However, although it is possible to calculate a number of different non-parametric effect sizes, it is not clear which non-parametric effect sizes should be used, nor how to aggregate results from individual experiments into an overall effect size.
7.6 Limitations
It should be noted that all primary studies using crossover designs (except Study 7 and Study 11), based their analysis on the pooled within treatment standard deviation, rather then the pooled within cell standard deviation. Both variances are calculated using a formula similar to that shown in (12) but the pooled within treatment variation is calculated based on pooling the variances of the observations in each of the two different treatment groups. In contrast, the pooled within cell standard deviation is based on pooling the variances calculated from the observations found in each of the experimental conditions shown in Table 8 for AB/BA crossover designs and Table 9 for 4-group crossover designs. This means the standard deviation will be biased (in fact the standard deviation will be larger than it should be), unless the system and period effects are negligible. Furthermore any bias in the standard deviation will impact the estimation of standardized effect size, making it smaller than it should be.
We claimed to have found a reproducibility problem if the difference between the effect size estimates reported by the authors and the ones we calculated was greater than 0.05. The choice of 0.05 was based on convenience and can be criticized. In practice, the value we chose seemed to work reasonably well as a means of drawing our attention to possible reproducibility problems. However, it incorrectly highlighted some differences that we believed to be due rounding errors, and we also observed two examples of accidental correctness. So, it was critical to review the actual meta-analysis process reported by the authors, as well as the difference between reported and calculated effect sizes to confirm whether there were validity or reproducibility problems.
8 Conclusions and Contributions
Our systematic review identified 13 primary studies from five high quality journals. In seven cases we identified validity or reproducibility problems. Even in cases where we reproduced the average standardized effect size, in four cases, we are not sure as to the accuracy of statistical tests of significance and p −values. We conclude that meta-analysis is not well understood by software engineering researchers.
Our systematic review process reported in Section 3 has ensured that the problems we identified were found in papers published in high quality software engineering journals with stringent peer review processes. It is, therefore, important to report such problems and provide guidelines and procedures to help to avoid them in the future. Answers to RQ1 and RQ2 reported in Section 4, provide traceability to the individual primary studies and contextual details of the experimental methods used to analyse each experiment. This confirms that we have not been biased in our selection of primary studies. Answers to RQ3 and RQ4 provide traceability to the individual meta-analysis problems and confirmation that most problems are found in more than one primary study, so are more than just one-off mistakes.
The major contributions of our study arise from our efforts to address the meta-analysis problems found by validity and reproduciblity assessment reported in Sections 5 and 6. They are:
- 1.
To provide evidence that meta-analysis methods are not well-understood by software engineering researchers (see Sections 5 and 6)
- 2.
To identify specific meta-analysis validity and reproducibility errors (see Sections 5 and 6).
- 3.
To provide guidelines for reporting and undertaking meta-analysis that could help to avoid meta-analysis errors (see Appendix A.6).
- 4.
To describe the model underlying the 4-group crossover experimental design (see Appendix A.1.3), since although the design is popular in software engineering research, it has not previously been specified in any detail.
- 5.
To provide a worked example of analyzing and meta-analyzing results from a family of studies that used a 4-group crossover design (see Appendix A.5).
Although we have provided meta-analysis reporting and conduct guidelines, it must be recognized that we lack the simulation studies needed to address questions such as:
Whether there is an optimum (or minimum viable) number of experiments in a family.
Whether the conversion to rpb is preferably to aggregating gIG directly, given the small sample sizes and numbers of independent experiments in SE families.
Whether we should use non-parametric methods for analysis and meta-analysis.
We are currently undertaking research addressing these issues.
Finally, whenever possible, we would ask researchers to make their data sets publicly available. Such data sets allow reviewers to check the validity of results before publication, provide a valuable resource for novice researchers, and allow data to be re-analyzed if new analysis methods become available.
Notes
Santos et al. (2018) reported that only 5 of the 39 papers they identified reported their raw data, so any reproducibility study we performed would need to be based primarily on summary statistics.
This criterion was amended after the protocol was completed because we identified the need to exclude correlation studies during data collection.
In our protocol we used the term correlation coefficient, however after beginning data extraction, we realized we needed to define the correlation coefficient effect size more correctly as the point-biserial correlation.
Since we were restricting ourselves to five international journals (see Section 3.2), we did not need to formally exclude extended abstracts or non-English papers.
Although Santos et al. (2018) found 15 families that used meta-analysis, three of the papers they found were excluded on the basis of our inclusion criteria and we found one study they did not.
That is, the same conceptual task e.g., fault detection, or a comprehension questionnaire, but with different materials (e.g., a different specification, design or code listing).
The tool is intended to help researchers aggregate experiments that use different design methods, and the between groups design is the most commonly used design method.
In the case of aggregated probability value there is no a priori value of P, so we can only make a subjective assessment of whether the calculated and reported values are close.
For example, for the metric Y.1 (Interest), the pretest score for group B was 0.81 and the posttest was 0.79 but the difference score was reported as − 0.03 (not − 0.02). This seems a minor issue, but since the difference score for group A was .1 and the pooled within group standard deviation of the difference score was 0.09. A difference score of − 0.03 for group B leads to an effect size of 1.444 while a difference score of − 0.02 leads to an effect size of 1.333 which after adjusting for the small sample size (nA = 5 and nB = 4) become 1.279 and 1.181 respectively.
Some papers reported forest plots with confidence bounds visible but it is not possible to extract accurate assessments of the values from such diagrams.
The standardised effect size variance is not the same as the sample variance. It is based on a formula including the number of participants in each different experimental condition and the standardised effect size itself.
Some researchers recommend using the standard deviation of the control group or the population standard deviation if it is known. See Lakens (2013) for a discussion of various different options for the choice of the standard deviation.
Please be aware that Hedges and Olkin called the unadjusted estimate of the standardized mean effect size g and the adjusted estimate d. Therefore, it is best to confirm explicitly whether or not the standardized mean effect size has been adjusted for small samples, rather that rely on using a possibly ambiguous label.
The following R code calculates J for numerical value x: sqrt(2/x)⋆gamma(x/2)/gamma((x-1)/2), and is easy to convert to a function.
In our reproducibility calculations we always used J(df).
This variance is not the same as the variance used to standardize the mean difference.
Heterogeneity is measured as an additional variance τ, which is added to the initial variance. The inverse of the revised variance is then used as the weight in the random effects meta-analysis. If τ is small, the effect on the meta-analysis results will be small.
Researchers wanting access to the data should contact Prof. Abrahão.
References
Abrahão S, Insfrán E, Carsí JA, Genero M (2011) Evaluating requirements modeling methods based on user perceptions: a family of experiments. Inf Sci 181 (16):3356–3378
Abrahao S, Gravino C, Insfran Pelozo E, Scanniello G, Tortora G (2013) Assessing the effectiveness of sequence diagrams in the comprehension of functional requirements: results from a family of five experiments. IEEE Trans Softw Eng 39 (3):327–342
Basili VR, Shull F, Lanubile F (1999) Building knowledge through families of experiments. IEEE Trans Softw Eng 25(4):456–473
BioStat (2006) Comprehensive meta-analysis (cma) v2.0. https://www.meta-analysis.com/pages/v2download.php
Borenstein M, Hedges LV, Higgins JPT, Rothstein HT (2009) Introduction to meta-analysis. Wiley, UK
Chow S, Liu J (1992) Design and analysis of bioavailability and bioequivalence studies. Taylor-Francis, New York
Ciolkowski M (2009) What do we know about perspective-based reading? An approach for quantitative aggregation in software engineering. In: Proceedings of the 2009 3rd international symposium on empirical software engineering and measurement. ESEM ’09. https://doi.org/10.1109/ESEM.2009.5316026. IEEE Computer Society, Washington, DC, pp 133–144
Cruz-Lemus JA, Genero M, Manso ME, Morasca S, Piattini M (2009) Assessing the understandability of UML statechart diagrams with composite states—a family of empirical studies. Empir Softw Eng 14(6):685–719
Cruz-Lemus JA, Genero M, Caivano D, Abrahão S, Insfrán E, Carsí JA (2011) Assessing the influence of stereotypes on the comprehension of UML sequence diagrams: a family of experiments. Inf Softw Technol 53(12):1391–1403
Cumming G (2012) Understanding the new statistics effect sizes, confidence intervals and meta-analysis. Routledge, UK
Dahl DB, Scott D, Roosen C, Magnusson A, Swinton J (2018) xtable: export tables to LaTeX or HTML. https://CRAN.R-project.org/package=xtable, r package version 1.8-3
Fernandez A, Abrahão S, Insfran E (2013) Empirical validation of a usability inspection method for model-driven Web development. J Syst Softw 86(1):161–186
Fernández-Sáez AM, Genero M, Chaudronand MRV, Caivano D, Ramos I (2015) Are forward design or reverse-engineered UML diagrams more helpful for code maintenance?: a family of experiments. Inf Softw Technol 57:644–663
Fernández-Sáez AM, Genero M, Caivano D, Chaudron MRV (2016) Does the level of detail of UML diagrams affect the maintainability of source code?: a family of experiments. Empir Softw Eng 21(1):212–259
Fisher R (1921) On the probable error of a coefficient of correlation deduced from a small sample. Metron 1:1–32
Gonzalez-Huerta J, Insfrán E, Abrahão S M, Scanniello G (2015) Validating a model-driven software architecture evaluation and improvement method: a family of experiments. Inf Softw Technol 57:405–429
Hadar I, Reinhartz-Berger I, Kuflik T, Perini A, Ricca F, Susi A (2013) Comparing the comprehensibility of requirements models expressed in Use Case and Tropos: results from a family of experiments. Inf Softw Technol 55(10):1823–1843
Hedges LV, Olkin I (1985) Statistical methods for meta-analysis. Academic Press, Orlando
Higgins JPT, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21(11):1539–1558
Hunter J, Schmidt F (1990) Methods of meta-analysis: correcting error and bias in research findings. Sage, Newbury Park
Johnson NL, Welch BL (1940) Applications of the non-central t-distribution. Biometrika 31(3–4):362–389
Jureczko M, Madeyski L (2015) Cross–project defect prediction with respect to code ownership model: an empirical study. e-Informatica Softw Eng J 9(1):21–35. https://doi.org/10.5277/e-Inf150102
Kitchenham B, Budgen D, Brereton P (2015) Evidence-based software engineering and systematic reviews. CRC Press, Boca Raton
Kitchenham B, Madeyski L, Budgen D, Keung J, Brereton P, Charters S, Gibbs S, Pohthong A (2017) Robust statistical methods for empirical software engineering. Empir Softw Eng 22(2):579–630
Kitchenham B, Madeyski L, Curtin F (2018) Corrections to effect size variances for continuous outcomes of cross-over clinical trials. Stat Med 37 (2):320–323. https://doi.org/10.1002/sim.7379. http://madeyski.e-informatyka.pl/download/KitchenhamMadeyskiCurtinSIM.pdf
Kitchenham B, Madeyski L, Brereton P (2019a) Problems with statistical practice in human-centric software engineering experiments. In: Proceedings of the 2019 international conference on evaluation and assessment in software engineering (EASE). https://doi.org/10.1145/3319008.3319009, pp 134–143
Kitchenham B, Madeyski L, Brereton P (2019b) Supplementary materials for the paper “Meta-analysis for families of experiments in software engineering: a systematic review and reproducibility and validity assessment”. http://madeyski.e-informatyka.pl/download/KitchenhamMadeyskiBrereton19Supplement.pdf
Laitenberger O, Emam KE, Harbich TG (2001) An internally replicated quasi-experimental comparison of checklist and perspective-based reading of code documents. IEEE Trans Softw Eng 27(5):387–418
Lakens D (2013) Calculating and reporting effect sizes to facilitate cumulative science: a practical primer for t-tests and anovas. Front Psychol 4(Article 863):1–12
Lipsey MW, Wilson DB (2001) Practical mete-analysis. Sage Publications Inc., UK
Madeyski L, Jureczko M (2015) Which process metrics can significantly improve defect prediction models? An empirical study. Softw Qual J 23(3):393–422. https://doi.org/10.1007/s11219-014-9241-7
Madeyski L, Kitchenham B (2017) Would wider adoption of reproducible research be beneficial for empirical software engineering research J Intell Fuzzy Syst 32(2):1509–1521. https://doi.org/10.3233/JIFS-169146. http://madeyski.e-informatyka.pl/download/MadeyskiKitchenham17JIFS.pdf
Madeyski L, Kitchenham B (2018a) Effect sizes and their variance for ab/ba crossover design studies. In: Proceedings of the ACM/IEEE 40th international conference on software engineering (May 27–June 3, 2018). ACM, Gothenburg, p 420, DOI https://doi.org/10.1145/3180155.3182556, (to appear in Print)
Madeyski L, Kitchenham BA (2018b) Effect sizes and their variance for AB/BA crossover design studies. Empir Softw Eng 23(4):1982–2017. https://doi.org/10.1007/s10664-017-9574-5
Madeyski L, Kitchenham B (2019) Reproducer: reproduce statistical analyses and meta-analyses. http://madeyski.e-informatyka.pl/reproducible-research/, R package version 0.3.0 (http://CRAN.R-project.org/package=reproducer)
Morales JM, Navarro E, Sánchez-Palma P, Alonso D (2016) A family of experiments to evaluate the understandability of TRiStar and i* for modeling teleo-reactive systems. J Syst Softw 114:82–100
Morris S B (2000) Distribution of the standardized mean change effect size for meta-analysis on repeated measures. Br J Math Stat Psychol 53:17–29
Morris S B, DeShon R P (2002) Combining effect size estimates in meta-analysis with repeated measures and independent-groups designs. Psychol Methods 7(1):105–125. https://doi.org/10.1037//1082-989X.7.1.105
Pfahl D, Laitenberger O, Ruhe G, Dorsch J, Krivobokova T (2004) Evaluating the learning effectiveness of using simulations in software project management education: results from a twice replicated experiment. Inf Softw Technol 46(2):127–147
Rosenthal R (1991) Meta-analytic procedures for social research. Sage, UK
Santos A, Gómez OS, Juristo N (2018) Analyzing families of experiments in SE: a systematic mapping study. CoRR arXiv:1805.09009
Scanniello G, Gravino C, Genero M, Cruz-Lemus J A, Tortora G (2014) On the impact of UML analysis models on source-code comprehensibility and modifiability. ACM Trans Softw Eng Methodol 23(2):13:1–13:26. https://doi.org/10.1145/2491912
Senn S (2002) Cross-over trials in clinical research, 2nd edn. Wiley, UK
Teruel MA, Navarro E, López-Jaquero V, Montero F, Jaen J, González P (2012) Analyzing the understandability of requirements engineering languages for CSCW systems: a family of experiments. Inf Softw Technol 54(11):1215–1228
Vegas S, Apa C, Juristo N (2016) Crossover designs in software engineering experiments: benefits and perils. IEEE Trans Softw Eng 42(2):120–135. https://doi.org/10.1109/TSE.2015.2467378
Viechtbauer W (2010) Conducting meta-analysis in R with the metafor package. J Stat Softw 36(3):1–48
Acknowledgements
We thank Silvia Abrahão, Carmine Gravino, Emilio Insfran, Guiseppe Scaniello and Genoveffa Tortora for for giving us access to their raw data. We are particularly grateful to Prof. Abrahão for providing us with details of her statistical analysis. We thank the reviewers for their helpful comments, particularly pointing out the issue of validity and the problem of aggregating invalid data. Lech Madeyski was partially supported by the Polish Ministry of Science and Higher Education under Wroclaw University of Science and Technology Grant 0401/0201/18.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: Jeffrey C. Carver
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A: Additional Statistical Details
Appendix A: Additional Statistical Details
This appendix provides additional statistical details that support the main paper.
1.1 A.1 Experimental Designs Used in the Primary Studies
In this section we describe the four different experimental designs used by our primary studies.
1.1.1 A.1.1 Independent Groups Design
The independent groups design, also referred to as a between-participants design, is the classic experimental design, where participants are randomly allocated to two groups. Participants in one group (group A) use one technique (with associated materials) to perform a task, and participants in the other group (group B) use the other technique (with the same materials) to perform the same task.
The standardized mean effect size (δIG, where IG stands for independent groups) is estimated by dividing the difference between the mean outcome for participants in group A and the mean outcome for participants in group B by the pooled within group standard deviation (see Lipsey and Wilson 2001; Borenstein et al. 2009, Hedges and Olkin 1985),Footnote 14 i.e.
where dIG is an estimate of δIG, MA is the mean value for participants in group A, MB is the mean value for participants in group B, and s is the pooled within group standard deviation, which is the square root of the pooled within group variance shown in (12).
where nA and nB refer to the number of observations in groups A and B respectively and varA and varB to the variance of the observations in groups A and B. If nA = nB, the pooled within group variance is simply the mean of varA and varB.
Equation (11) makes it clear that effect sizes have direction as well as magnitude. Researchers aggregating results from a family of experiments must ensure that all effect sizes adopt the same direction for the difference. This is straightforward if there is a well-defined control method, otherwise the decision is arbitrary but must be consistent.
Equation (11) is a valid estimate of the standardized difference between Technique A and Technique B. However, for small sample sizes, the estimate is biased and should be corrected, as recommended by Hedges and Olkin (1985), to give an improved estimate:Footnote 15
J(df) is calculated from the formula:Footnote 16
where Γ is the Gamma distribution and the degrees of freedom (df ) is the number of participants minus 2 (because of the two groups). J tends to 1 as the sample size increases, so rather than apply some arbitrary cutoff point to stop applying the correction, it is sensible to always apply it whatever the sample size. J(df) is often approximated by c(df) for sample sizes greater than 10 using the formula:Footnote 17
Most meta-analyst researchers recommend aggregating the standardized effect sizes using a weighted average, where the weights are based on the inverse of the variance of the standardized effect size (see Borenstein et al. 2009 or Lipsey and Wilson2001).Footnote 18 The normal approximation to the exact formula for the estimate of a standardized effect variance of δIG is reported in Borenstein et al. (2009):
here nA is the number of participants in group A and nB is the number of participants in group B. It should be noted that this equation is inaccurate for very small sample sizes (Morris 2000).
In order to find the variance of gIG, multiply the right-hand side of (16) by [J(df)]2 and let \([J(df)]^{2}d^{2}_{IG}=g^{2}_{IG}\):
If nA = nB = n and we let 2n = N:
This is the same formula used by Pfahl et al. (2004) to find the variance of their standardized effect size (see Appendix B in (Pfahl et al. 2004)) which they used both to perform homogeneity tests and to calculate the overall weighted average. To test for homogeneity, Pfahl et al. (2004) used Q as proposed by Hedges and Olkin (1985):
where \(Var(d_{IG})=\hat {\sigma }(d_{i})^{2}\).
Although the above discussion might appear quite complex, the independent groups design is the most straightforward experimental design to meta-analyze using a mean difference effect size.
1.1.2 A.1.2 AB/BA Crossover Design
The AB/BA Crossover design (see Senn 2002; Vegas et al. 2016; and Madeyski and Kitchenham 2018a, b) is a repeated measures design which was used by four families. In an AB/BA crossover, participants are spilt into two groups and each group uses one of the competing techniques with one set of materials. Subsequently, they perform the same task with a second set of materials, with each group using the other technique. The design is illustrated in Table 8.
The details of this analysis for the standard AB/BA crossover design can be found in Madeyski and Kitchenham (2018b). As discussed in Section 4, all crossover designs have two different types of standardized mean difference effect size, δRM estimated by dRM using (1) and δIG estimated by dIG using (2).
Equation (1) is a valid estimate of the standardized difference between Technique A and Technique B assuming that there is no significant technique by period interaction. For small sample sizes, the estimate is biased and should be multiplied by J(df) to give an improved estimate (Hedges and Olkin 1985):
where the degrees of freedom (df ) is the number of participants minus 2 (because of the two sequence groups). It is extremely important to note that the degrees of freedom relate to the number participants not the number of observations. We explain the reason for this below.
Because gRM is an unbiased estimate of the unstandardized mean difference divided by its variance, the equation for the t −test value related to δRM is:
where nA and nB are the number of observations in group A and group B respectively. As pointed out by Madeyski and Kitchenham (2018b), because the exact variance of a t −variable is known (Johnson and Welch 1940), the variance of gRM can be calculated by multiplying the formula for the variance of a t −variable by \({(\frac {1}{n_{A}} + \frac {1}{n_{B}})}\).
gIG can be calculated from the relationship between gRM and gIG, see (4). It can also be calculated directly from the raw data. dIG is based on the standardized mean difference, using sIG as the standardizer. We can estimate sIG by pooling the within cell variance for each of the four cells in Table 8 (although this assumes that the variance of each cell is estimating the same population variance). This is because with any cell, the conditions (i.e., technique, time period, material used) are the same for all participants whose results are in that cell. As pointed out by Madeyski and Kitchenham (2018b), if we assume that each condition is represented as a numerical effect, then each participant in a cell is modelled by the formula:
where yi is the i th participant in the cell, μi is the mean for subject yi, Tj, Pk and Ml are the effects for technique j, time period k and materials l, respectively, and ei is an error term assumed to be normally distributed with zero mean and variance \(s^{2}_{IG}\). Standard statistical theory says that var(x) = var(x + A) where A is any constant. So if μi, Tj, Pk and Ml are assumed to be constants, the variance of the yi-values is an unbiased estimate of \(s^{2}_{IG}\). Assuming a single population variance, pooling the data from all four cell should provide a more precise estimate of \(s^{2}_{IG}\) than would be obtained by pooling only the cells in the first time period.
However, if we mix up the data from two cells, for example, in the context of an AB/BA crossover, if we put the observations that used technique T1 together, we have some subjects with the model:
and others with the model:
Then, unless, P1 + M1 = P2 + M2, calculating the variance of the data from the two combined cells will not result in an unbiased estimate of \(s^{2}_{IG}\). The differences between the time period and material effects will inflate the estimate of the variance. This is, of course, the theory underlying fixed effects analysis of variance.
Furthermore, although, the repeated measures allow us to calculate \(s^{2}_{IG}\) with increased precision, if we have only N participants, our estimates are based on the variation among those N participants. No matter how many times we take repeated measures on those N participants, the degrees of freedom relating to the variance remain the same, because our estimate of the population variance is still based on the same N participants.
1.1.3 A.1.3 4-Group AB/BA Crossover Design
The 4-group AB/BA cross over design is a variant of the AB/BA crossover, where the basic design is duplicated with the materials used in period one and the materials used in period two exchanged. The design is illustrated in Table 9. The design appears to be unique to software engineering studiesFootnote 19 and was used by seven families.
Like the standard AB/BA crossover, this design permits researchers to calculate both a repeated measures effect size and an effect size equivalent to an independent groups effect size. Comparing Tables 8 and 9, it is clear that the 4-group crossover is based on two balanced AB/BA crossovers that differ only in the order in which the materials are used. Groups A and B correspond to the one AB/BA crossover while Groups C and D correspond to the other.
The design can be understood by considering the impact on a participant in each of the four groups and in each time period. We developed a model of the 4-group crossover that is shown in Table 10. The terms identify the conditions and outcome value for each participant in each cell:
- 1.
yg,h,i identifies the outcome measure for for participant i in time period h = 1, 2 using technique g = 1, 2.
- 2.
μi is the average outcome measure for participant j
- 3.
τg is the effect for technique g
- 4.
Mf where f = 1, 2 is the effect of performing the required task using one of the two different software applications (as represented by each application’s specifications, code, documents etc.)
- 5.
π is any systematic effect resulting from doing the same task a second time.
- 6.
COx where x = 1, 2 identifies which of the two duplicated crossovers a participant belongs to.
- 7.
λq where q = 1, 2 is the effect of performing the task for a second time using one technique, after first performing the task using the other technique. The value of q specifies which technique was used first. Following the advice of Senn (2002) for simple AB/BA crossovers, we assume that λq = 0, and all other possible interactions are likewise zero.
Analysis of the 4 group crossover can be understood by subtracting the outcome from time period P1 from the outcome from time period P2. This assumes that the outcome is a suitable measure, such as a measure of the time to complete a task. For measures related to understandability, the number of correct answers is acceptable unless the values are very restricted (i.e., the number of correct out of 10 is acceptable, the number correct out of two is not). The effect of calculating the time period difference is shown in Table 11. The impact of calculating the difference is to remove the effect due to the individual participant.
If we take the average of the difference values in group (i.e., calculate \(\overline {DI}\) where I = 1,..., 4), it is easy to see that, in terms of expected values, we have:
where τ2 − τ1 is the unstandardized effect size. In fact, the unstandardized effect size can also be calculated by subtracting the mean value of all observations derived from participants using technique T1 from the mean value of all observations derived from participants using technique T2. However, the formal model underlying each cell makes it clear that in order to estimate the between participants variance \(s^{2}_{IG}\), it is necessary to construct the pooled within cell variance. Using the pooled variance of all observations derived from participants using the same technique would inflate the variance because subsets of the data points would be affected by different factors.
We provide a brief tutorial on analyzing and meta-analysizing data from 4-group crossover designs in Appendix A.5.
1.1.4 A.1.4 Pretest Posttest Control Group Design
The pretest posttest control group design is a repeated measures design, but rather different from a crossover style design. In this design, participants are randomly allocated to two groups. Then, both groups undertake the same test (or perform the same SE activity) using their current technique. The groups are then split and participants in one group receive one type of training and participants in the other group are given a competing form of training. They are then asked to undertake another test. This design is illustrated in Table 12. It was used only in Study 14. It is not necessary for the pretest and posttest tasks to be the same. However, in Study 14, the authors asked participants to undertake a test on their SE knowledge and repeated the same test after their training.
Although, this is a repeated measures design it has rather different properties to a crossover style design. In fact, if analysts work solely with the difference scores, the data can be analysed as if the difference data were the outcome of an independent groups study. This form of analysis is called a difference of differences analysis and the standardised effect size measures the relative difference in the average individual improvement of participants in group A compared with participants in group B.
This design includes one of the main advantages of a crossover design that is, the effect of individual differences are catered for by the analysis, but avoids the problem of technique by period interaction which is a potential risk when using a crossover design. For example, there were many studies evaluating the perspective-based code reading (PBR) methods (Ciolkowski 2009), some of which used the undefined current method as a control while others used the checklist-based reading (CBR) method as a control. Using a pretest posttest control group, the current method would be used to establish a pretest baseline and then groups could be randomly assigned to training in CBR or PBR and the posttest differences used to assess whether PBR or CBR most enhanced defect detection.
A model of the experimental design for each cell and for the difference data is shown in Table 13.
The model assumes a situation such as we discussed above for code reading methods, when there are three treatment conditions, one control that is used before training and then half the participants receive training in one alternative treatment and the other half receive training in the other. The effect of subtracting the mean difference values of group A from the mean difference values of group B is to obtain an estimate of τ1 − τ2 which is the unstandardized effect size. The basic design can easily be revised to cater for only two conditions (i.e., control and treatment conditions) by letting all subjects use the control conditions in time period 1 and in time period 2 to let participants in Group A use the treatment and participants in Group B to use the control. The difference between the difference values then equates to τ1 − τc. Effect size construction and the effect size variance formulas for this design are discussed in Morris and DeShon (2002).
This design can also be used if the pretest does not involve performing the same tasks that is done in the posstest. This method allows for situations where participant skill is measured by some other means (e.g., the results of a completely different software engineering task, or, for students, their previous year grades). In this version of the design the pretest values for each participants are used as a covariate in an ANCOVA analysis.
1.2 A.2 Meta-analysis Based on the Relationship Between the Standardized Mean Difference and the Point Bi-serial Correlation
Like any correlation, rpb is the correlation between two values x and y. However, for rpb, y is the value of the outcome metric and x is a categorical variable taking the value zero if y was obtained from a participant in the control group and one if y was obtained from a participant in the treatment group. Clearly, rpb is not a valid Pearson correlation coefficient because it is not the correlation between two normally distributed variables, and it is often referred to as a pseudo-correlation. In practice, rpb is often calculated as the square root of the multiple correlation coefficient, R2, which in the context of a one-way ANOVA is calculated as the percentage reduction in the total variation due to removing the between group variation. The danger with calculating rpb from R2 is that the direction of the effect is lost.
The process to convert from a standardized mean effect size, derived from descriptive statistics, to a point bi-serial correlation effect size is as follows:
- 1.
For each individual experiment in a family, estimate dIG from the difference between the mean values for each technique group and pooled within technique group standard deviation. Then apply the small sample size adjustment factor based on the number of participants to calculate gIG.
- 2.
Converte gIG to the point bi-serial correlation rpb using the formula:
$$ r_{pb}=\frac{g_{IG}}{\sqrt{g_{IG}^{2}+a}} $$(26)where a = (nA + nB)2/(nAnB) and a = 4 if nA = nB (see Borenstein et al. 2009). For AB/BA crossover designs (both standard crossover and the 4-group crossover), nA is the number of participants that used technique A in period 1 and nB is the number of participants that used technique B in period 1.
- 3.
Apply the Fisher normalisation formula (Fisher 1921) to the rpb values for each experiment:
$$ Zr=0.5\frac{ln(1+r_{pb})}{ln(1-r_{pb})} $$(27)and the variance of each Zr is:
$$ var(Zr)=\frac{1}{(n_{A}+n_{B}-3)} $$(28) - 4.
Use the R metafor library to perform meta-analysis on Zr. Assuming a fixed effects model, the aggregate value of Zri for a family of experiments is calculated from the formula:
$$ \overline{Z}_{r}=\frac{{\sum}_{i} w_{i}Zr_{i}}{{\sum}_{i} w_{i}} $$(29)where wi = 1/var(Zri) = nA + nB − 3 and i is the i th experiment in the family. The variance of \(\overline {Z}_{r}\) is calculated from the formula:
$$ var(\overline{Z_{r}})=\frac{1}{{\sum}_{i} w_{i}} $$(30)Although such formulas can easily be applied manually, metafor is useful for calculating confidence intervals and producing forest plots. It also allows meta-analysts to perform a random effects analysis. A priori, a fixed effects analysis should be reasonable for families of experiments, when the different experiments in a family all test the same hypotheses, and use both the same experimental designs and the same materials. Table 1 in the supplementary material (Kitchenham et al. 2019b) reports the differences among experiments in each family. From that table, it appears that a random effects model might be preferable only for Study 1 and Study 3. However, applying a random effects analysis when there is no significant heterogeneity among studies gives results very similar to a fixed effects analysis.Footnote 20 Thus, we recommend using a random effects for all analyses in order to check whether there is a substantial level of heterogeneity.
- 5.
Results in the transformed Zr scale need to be back transformed first to rpb and then to gIG. For example to convert back to the weighted mean of the gIG values, the following two transformations are needed:
$$ \overline{r_{pb}}=\frac{e^{2\overline{Z_{r}}}-1}{e^{2\overline{Z_{r}}}+1} $$(31)and
$$ \overline{g_{IG}}=\overline{r_{pb}}\sqrt{\frac{a}{(1-\overline{r_{pb}}^{2})}} $$(32)where a = (nA + nB)2/(nAnB) and a = 4 if nA = nB.
1.3 A.3 Meta-analysis Using the Point Bi-serial Correlation and the Hunter Schmidt Method
Study 8 reported rpb and used it in their meta-analysis. However, they did not derive rpb from a standardized effect size, but from the one-sided probabilities of significance from the hypothesis tests for each experiment, i.e., the p-values. For each experiment in the family and for each metric, they used the p − value obtained either the Mann-Whitney-Wilcoxon (MMW) test or the t −test depending on the outcome of a normality test.
The p −values must come from one-sided tests in order to preserve the direction of the effect size. For example, if we are testing whether method A is more efficient that method B, a large one-sided probability (e.g., 0.96) would give a z-value of 1.751 and would indicate that method A was more efficient that method B. A small one-sided probability (e.g., 0.04) would give a z-value of − 1.751 and indicate method B was more efficient than method A.
The authors of Study 8 report using the equation:
This is not ideal because it does not make it clear that rpb can potentially be negative.
The study authors used the Hunter-Schmidt method to aggregate their correlations:
Then, the variance of \(\bar {r}\) is given by the equation:
They, also, appear to have used the number of observations as the basis for ni rather than the number of participants. This is because the authors report that their family included 92 participants, but report N = 184 for their overall mean \(\bar {r}\). However, in this case using 2ni rather than ni in (34) to calculate the variance of \(\bar {r}\) has no effect on the value, because two is a multiplicative constant in both the top and bottom of the fraction and cancels out. The only equation that is affected by using the wrong sample size is the formula for χ2 that is used to test heterogeneity:
The effect of using 2ni rather than ni in (36) is to quadruple the value of χ2 and increase the likelihood of incorrectly assuming that the effect sizes were heterogeneous.
1.4 A.4 Aggregating p −values
Both Study 13 and Study 14 aggregated one-sided p −values in order to test the null hypothesis of no significant difference between techniques. They tested whether the p −values were heterogeneous using the equation:
where, under homogeneity, Q is χ2 with k − 1 degrees of freedom, and zi is the standard normal deviate corresponding to the one-tailed p −values.
Then, they aggregated the p −values using the formula:
They tested whether P was significant using the χ2 distribution with 2k degrees of freedom. This approach, which is sometimes called Fisher’s method, has a number of important limitations, particularly if the p −values exhibit heterogeneity, and is no longer recommended (Rosenthal 1991).
1.5 A.5 Parametric Analysis and Meta-analysis of Crossover Design Experiments
In this section, we provide guidelines for analyzing and meta-analyzing crossover style experiments. In particular, we provide an example of analyzing the 4-group using the data provided by Prof. Abrahão.Footnote 21
We use the R linear mixed model package lme4 to analyze data from individual experiments. In the case of a conventional two group AB/BA crossover, for each experiment, we use a model including fixed effects:
Time Period with values P1 and P2.
Technique with values T1 and T2.
The personal identifier for each person is treated as a random effects factor. An example of this analysis, explaining how to obtain the estimates of dIG and dRM can be be found in Madeyski and Kitchenham (2018b). The data is held in what is referred to as the long format, that is there are two entries for each participant that define the conditions under which each outcome observation was obtained.
To analyze a 4-group AB/BA crossover We used a model that included fixed effects factors specifying:
Time Period with values P1 and P2.
Technique being compared with values T1 and T2.
The Objects (i.e., software materials) being used with values determined by the names given to the software object being investigated.
The crossover duplicate pair to which the participant belonged which had values COD1 which refers to a participant in Group A or Group B and COD2 which refers to a participant in Group C or Group D. The crossover pair factor identifies the groups that used materials in the same order.
Participant identifier (“ID”) was used as the random effects factor. Using this model with Prof. Abrahão’s data from her Italy1 experiment, we obtained the analysis shown in Fig. 1. Assuming the data are held in a data frame called Italy1 (in a format corresponding to the hypothetical data shown in Table 14 which reports the data for four participants), the R instructions to perform this analysis are presented in Output 1:

The assumptions underlying this analysis are:
- 1.
All observations are normally distributed.
- 2.
Variances calculated from each cell are all estimating the same underlying population variance.
There are several things to note:
- 1.
The analysis constructs a name for fixed effect sizes based on the name of the categorical variable and the label(s) given to categorical values. The label name used is the second in alphabetical order. So since labels for the Method variable are NODM and DM, the package calculates the effect size as NODM − DM. This is why the value of the Method effect size is negative. Since we consider the DM condition to be the treatment condition and the NODM condition to be the control, we define the unstandardized treatment effect to be − NoDM = .02125.
- 2.
The estimate of the within participant variance is given by the Random Effects residual term.
- 3.
COD2 corresponds to the fixed effect size of the difference between results for the A and B crossover and the results for the C and D crossover. Since the difference between the groups is the order in which they used the Objects (i.e., the application specifications), they indicate that documents related to EPlat were more difficult to understand than documents related to the other specification (ECP).
- 4.
The variance associated with the random effects ID terms is the estimate of the between participants variance.
- 5.
The standard error of the COD2 fixed effect size is larger than the other fixed effect sizes. This is because it is based on the between participants variance.
The estimate of the variance of an individual participant observation is the sum of the between subjects and within subjects variance i.e., \(s^{2}_{IG}\). In the case of the Italy1 data set we have the estimate of \(s^{2}_{IG}\) taking the value 0.01863 + 0.01029 = 0.02892.
Then, from the linear mixed model analysis
The estimate of dRM is \(0.02125/\sqrt {0.01029}=0.2095 \).
The estimate of dIG is \(0.02125/\sqrt {0.02892}= 0.125\).
The estimate of r, the correlation between repeated measures is r = 0.01863/(0.01863 + 0.01029) = 0.6442
Using this method, we obtained standardized effect sizes and the correlation between participants for each of the five experiments undertaken by Prof. Abrahão’s and her colleagues. These results are shown in Table 15.
We applied the exact small sample size adjustment to dIG and dRM. We used (26) to calculate equivalent point bi-serial correlation effect sizes and applied Fisher’s normalising transformation to obtain the zRM and zIG values. The variances for the zRM and zIG values are calculated as v(z) = 1/(ni − 3) (which is the same for both variables from the same experiment). These results are shown in Table 16. The results for gIG obtained for each experiment are quite close to, but slightly larger than, the ones we obtained using the published descriptive statistics reported in Table 5. This is because we have fitted a more complex model to the data that accounts for all the built-in blocking factors in the experimental design and, so, provides us with a more accurate estimate of the between participant variance. When blocking factors have a significant effect on the experiment outcomes, we would expect variance estimates from the full model to be smaller than those from the descriptive statistics, so the effect size estimates should be larger. The gRM values are larger than the gIG values because of the correlation between the repeated measures.
We used the metafor package to analyze the zIG and zRM data. For example, to analyse the zIG data we used the R instructions in Output 2:

This produced the meta-analysis results summarized in Fig. 2. These results are still in the transformed data scale. Figure 3 shows a forest plot of the meta-analysis results transformed back to the gIG scale.
Assuming the meta-analysis results from the rma function call are saved into a R data structure labelled AbrahaoResults, the R instructions needed to report the contents of Fig. 3 as a pdf file are:

The parameter transformZrtoHgappro identifies a function we created in order for the forest function to transform from the normalized point bi-serial correlation back to the corresponding standardized mean difference effect size. The function is only permitted to have one parameter (a value corresponding to a transformed point bi-serial correlation), which means that we must assume a balanced experiment because we cannot include different group sizes as parameters, i.e. the function assumes that there are the same number of participants in groups A and B as there are in groups C and D. If this is not the case the forest plot values will be slightly biased. The instruction text is used to annotate the forest plot. In our experience the actual values required to put the annotations in the correct places need to be determined by trial and error.
The meta-analysis results for gIG and gRM are summarized in Table 17. These have been transformed to the standardized mean different effect size using functions that allow for unbalanced experiments. The functions we use to transform between the effect sizes are available in our Reproducer package (see Appendix A.7).
The p −value for gIG is less than the p −value for gRM because there is significant heterogeneity among the gRM effect sizes (QEp = 0.033). This means that the standard error of the mean is increased for gRM. The confidence interval bounds on the overall mean gRM are wider than the confidence limits bounds on gIG for the same reason.
1.6 A.6 Guidelines for Meta-analysis Reporting and Practice
After analysing the reporting and conduct of our primary studies, we recommend the following reporting guidelines:
Use sufficient precision to report descriptive statistics, in terms of the number of decimal points used to report data.
Report the values of descriptive statistics not only figures such as box plots. It is preferable to include both the actual values and the graphical displays.
For repeated measures designs, report the correlation between the repeated measures.
Specify the particular version of the standardized mean difference effect size using a formula rather than a name.
Confirm whether or not the small sample size adjustment has been applied to any reported standardized mean difference effect sizes.
Specify the model used to aggregate the experiment effect sizes, i.e., fixed, random or mixed.
Report the results of the aggregation process including the overall effect size, its p −value, and confidence limit bounds, the heterogeneity test statistic (Q) and its p −value. In the case of relatively large heterogeneity, it is also worth reporting the estimate of the heterogeneity statistic.
With respect to performing meta-analysis, our results suggest researchers:
Should understand the implication of the design of each experiment on effect sizes and their variances.
Should ensure that effect sizes obtained from experiments that used different designs are equivalent.
Should be careful to maintain the direction as well as the magnitude of effect sizes. When meta-analyzing effect sizes, all the effect sizes must be based on investigating whether the effect of one specific technique is greater than the effect of the other technique, and must allow the effect size to be positive or negative. This includes occasions where the effect sizes are derived from the one-sided p −values.
Should undertake sanity checks of the outcomes from meta-analysis tools based on their descriptive statistics.
Should use a random effects model for aggregating effect sizes, unless there is a very strong argument for using a fixed effects model.
Should be careful about using general purpose meta-analysis tools. General purpose tools are designed to handle a variety of different experimental designs by converting the results from complex designs to the simplest design (i.e., an independent groups analysis). However, to support multiple experimental designs, they may have complex interfaces. In addition, they may not support newly developed experimental designs.
1.7 A.7 Reproducibility of this Paper
To support the reproducibility of this paper, it is complemented by the reproducer R package (Madeyski and Kitchenham 2019) (available from CRAN—the official repository of R packages). The reproducer package includes both the collected data sets from the analyzed studies and the computational procedures developed by the first two authors (e.g., calculateSmallSampleSizeAdjustment, constructEffectSizes, transformRtoZr, transformZrtoR, transformHgtoR, calculateHg, transformRtoHg, transformZrtoHgapprox, transformZrtoHg) that are used to reproduce the results (e.g., Tables 5, 6, and 7 were automatically generated on a basis of the collected data sets and functions included in reproducer). Our aim is to promote reproducibility of research in empirical software engineering (Madeyski and Kitchenham 2017) by supporting our research papers by the related R package (see Madeyski and Kitchenham 2018b; Kitchenham et al. 2017; Jureczko and Madeyski 2015; Madeyski and Jureczko 2015).
In Madeyski and Kitchenham (2017) we emphasized that reproducible research (RR) refers to the idea that the ultimate product of research is the paper plus its computational environment. Therefore, our RR document that incorporates the textual body of the paper and calls to the reproducer R functions including analysis steps (e.g., functions to calculate and transform different effect sizes) used to process the data, as well as calls to the xtable R package (Dahl et al. 2018) that helps us to automatically present results in a tabular form will be available upon request from the corresponding author for reviewers and researchers interested in building on the outcomes presented in the paper. This RR document along with reproducer available in R environment can be used to compile all pieces of information into the resulting document in the pdf format.
An important part of documenting the research process with R is recording the R session info, which makes it easier for future researchers to recreate what was done in the past and which versions of the R packages were used. The information from the session we used to create this paper is shown in Output 4:

Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kitchenham, B., Madeyski, L. & Brereton, P. Meta-analysis for families of experiments in software engineering: a systematic review and reproducibility and validity assessment. Empir Software Eng 25, 353–401 (2020). https://doi.org/10.1007/s10664-019-09747-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10664-019-09747-0