Abstract
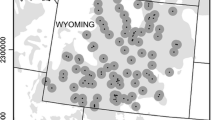
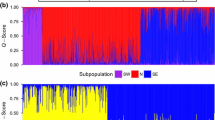
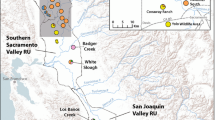
Greater sage-grouse (Centrocercus urophasianus) within the Bi-State Management Zone (area along the border between Nevada and California) are geographically isolated on the southwestern edge of the species’ range. Previous research demonstrated that this population is genetically unique, with a high proportion of unique mitochondrial DNA (mtDNA) haplotypes and with significant differences in microsatellite allele frequencies compared to populations across the species’ range. As a result, this population was considered a distinct population segment (DPS) and was recently proposed for listing as threatened under the U.S. Endangered Species Act. A more comprehensive understanding of the boundaries of this genetically unique population (where the Bi-State population begins) and an examination of genetic structure within the Bi-State is needed to help guide effective management decisions. We collected DNA from eight sampling locales within the Bi-State (N = 181) and compared those samples to previously collected DNA from the two most proximal populations outside of the Bi-State DPS, generating mtDNA sequence data and amplifying 15 nuclear microsatellites. Both mtDNA and microsatellite analyses support the idea that the Bi-State DPS represents a genetically unique population, which has likely been separated for thousands of years. Seven mtDNA haplotypes were found exclusively in the Bi-State population and represented 73 % of individuals, while three haplotypes were shared with neighboring populations. In the microsatellite analyses both STRUCTURE and FCA separate the Bi-State from the neighboring populations. We also found genetic structure within the Bi-State as both types of data revealed differences between the northern and southern part of the Bi-State and there was evidence of isolation-by-distance. STRUCTURE revealed three subpopulations within the Bi-State consisting of the northern Pine Nut Mountains (PNa), mid Bi-State, and White Mountains (WM) following a north–south gradient. This genetic subdivision within the Bi-State is likely the result of habitat loss and fragmentation that has been exacerbated by recent human activities and the encroachment of singleleaf pinyon (Pinus monophylla) and juniper (Juniperus spp.) trees. While genetic concerns may be only one of many priorities for the conservation and management of the Bi-State greater sage-grouse, we believe that they warrant attention along with other issues (e.g., quality of sagebrush habitat, preventing future loss of habitat). Management actions that promote genetic connectivity, especially with respect to WM and PNa, may be critical to the long-term viability of the Bi-State DPS.





Similar content being viewed by others
References
Balloux F, Lugon-Moulin N (2002) The estimation of population differentiation with microsatellite markers. Mol Ecol 11:155–165
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (2004) GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5171, Université de Montpellier II, Montpellier
Benedict NG, Oyler-McCance SJ, Taylor SE, Braun CE, Quinn TW (2003) Evaluation of the eastern (Centrocercus urophasianus urophasianus) and western (Centrocercus urophasianus phaios) sub-species of Sage-grouse using mitochondrial control region sequence data. Conserv Genet 4:301–310
Bi-State Technical Advisory Committee Nevada and California (2012) Bi-State action plan: past, present, and future actions for conservation of the greater sage-grouse Bi-State Distinct Population Segment. Prepared for the Bi-State Executive Oversight Committee for Conservation of Greater Sage-Grouse
Bonnet E, Van de Peer Y (2002) ZT: a software tool for simple and partial Mantel tests. J Stat Softw 7:1–12
Bouzat JL, Cheng HH, Lewin HA, Westemeier RL, Brawn JD, Paige KN (1998) Genetic evaluation of a demographic bottleneck in the greater prairie chicken. Conserv Biol 12:836–843
Braun CE (1998) Sage grouse declines in western North America: what are the problems? Proc West Assoc State Fish Wildl Agencies 78:139–156
Caizergues A, Dubois S, Mondor G, Loiseau A, Ellison LN, Rasplus JY (2001) Genetic structure of black grouse (Tetrao tetrix) populations of the French Alps. Genet Sel Evol 33:S177–S191
Caizergues A, Rätti O, Helle P, Rotelli L, Ellison L, Rasplus JY (2003) Population genetic structure of male black grouse (Tetrao tetrix L.) in fragmented vs. continuous landscapes. Mol Ecol 12:2297–2305
Casazza ML, Coates PS, Overton CT (2011) Linking habitat selection and brood success in greater sage-grouse. In: Sandercock BK, Martin K, Segelbacher E (eds) Ecology, conservation, and management of grouse. Studies in Avian Biology, vol 39. University of California Press, Berkeley, pp 151–167
Commons ML, Baydack RK, Braun CE (1999) Sage grouse response to pinyon–juniper management. In: Monson SB, Stevens R (eds) Ecology and management of pinyon–juniper communities within the interior West. USDA Forest Service RMRS-P-9, USDA Forest Service Rocky Mountain Research Station, Ft. Collins, pp 238–239
Connelly JW, Braun CE, Knick ST, Baker WL, Beever EA, Christiansen TJ, Doherty KE, Garton EA, Hagen CA, Hanser SE, Johnson DH, Leu M, Miller RF, Naugle DE, Oyler-McCance SJ, Pyke DA, Reese KP, Schroeder MA, Stiver SJ, Walker BL, Wisdom MJ (2011) Conservation of greater sage-grouse: a synthesis of current trends and future management. In: Knick ST, Connelly JW (eds) Greater sage-grouse: ecology and conservation of a landscape species and its habitats, vol 38., Studies in Avian Biology SeriesUniversity of California Press, Berkley, pp 549–564
Earl D, vonHolt BM (2011) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. doi:10.1007/s12686-011-9548-7
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Giesen KM, Schoenberg TJ, Braun CE (1982) Methods for trapping sage grouse in Colorado. Wildl Soc Bull 10:224–231
Goudet J (1995) Fstat version 1.2: a computer program to calculate F statistics. J Hered 86:485–486
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Guo SW, Thompson EA (1992) Performing the exact test of Hardy–Weinberg proportions for multiple alleles. Biometrics 48:361–372
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Johnsgard PA (1983) The grouse of the world. University of Nebraska Press, Lincoln
Kahn NW, Braun CE, Young JR, Wood S, Mata DR, Quinn TW (1999) Molecular analysis of genetic variation among large and small-bodied sage grouse using mitochondrial control-region sequences. Auk 116:819–824
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Mooney HA, St. Andre G, Wright RD (1962) Alpine and subalpine vegetation patterns in the White Mountains of California. Am Midl Nat 68:257–273
Naugle DE, Aldridge CL, Walker BL, Cornish TE, Moynahan BJ, Holloran MJ et al (2004) West Nile virus: pending crisis for greater sage-grouse. Ecol Lett 7:704–713
O’Brien SJ, Evermann JF (1988) Interactive influence of infectious disease on genetic diversity of natural populations. Trends Ecol Evol 3:254–259
Oyler-McCance SJ, St. John J (2010) Characterization of small microsatellite loci for use in non invasive sampling studies of Gunnison sage-grouse (Centrocercus minimus). Conserv Genet Res 2:17–20
Oyler-McCance SJ, Taylor SE, Quinn TW (2005a) A multilocus population genetic survey of the greater sage-grouse across their range. Mol Ecol 14:1293–1310
Oyler-McCance SJ, St. John J, Taylor SE, Apa AD, Quinn TW (2005b) Population genetics of Gunnison sage-grouse: implications for management. J Wildl Manag 69:630–637
Oyler-McCance SJ, St. John J, Quinn TW (2010) Rapid evolution in lekking grouse: implications for taxonomic definitions. Ornithol Monogr 67:114–122
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Piertney SB, Höglund J (2001) Polymorphic microsatellite DNA markers in black grouse (Tetrao tetrix). Mol Ecol Notes 1:303–304
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256
Pritchard JK, Stephens M, Donnelly PJ (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pritchard JK, Wen X, Falusch D (2010) Documentation for STRUCTURE software: version 2.3. http://pritchardlab.stanford.edu/structure_software/release_versions/v2.3.4/structure_doc.pdf. Accessed 15 Oct 2013
Quattro JM, Vrijenhoek RC (1989) Fitness differences among remnant populations of the endangered Sonoran topminnow. Science 245:976–978
Quinn TW (1992) The genetic legacy of Mother Goose—phylogeographic patterns of lesser snow goose Chen caerulescens caerulescens maternal lineages. Mol Ecol 1:105–117
Quinn TW, Wilson AC (1993) Sequence evolution in and around the mitochondrial control region in birds. J Mol Evol 37:417–425
Rambaut A (2008) FigTree: tree figure drawing tool, version 1.3.1. http://tree.bio.ed.ac.uk/software/figtree/. Accessed 1 Aug 2012
Raymond M, Rousset M (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Rosenberg NA (2002) Distruct: a program for the graphical display of structure results. http://www.cmb.usc.edu/~noahr/distruct.html
Schneider S, Roessli D, Excoffier L (2001) Arlequin: a software for population genetics data analysis. Version 2.001. Genetics and Biometry Laboratory, University of Geneva, Geneva
Schroeder MA (2008) Variation in greater sage-grouse morphology by region and population: U.S. Fish and Wildlife Service, unpublished report
Schroeder MA, Aldridge CL, Apa AD, Bohne JR, Braun CE et al (2004) Distribution of sage grouse in North America. Condor 106:363–376
Segelbacher G, Paxton RJ, Steinbrück G, Tronteljj P, Storch I (2000) Characterization of microsatellites in capercaillie Tetrao urogallus (AVES). Mol Ecol 9:1934–1935
Spaulding A (2007) Rapid courtship evolution in grouse (Tetraonidae): contrasting patterns of acceleration between Eurasian and North American polygynous clades. Proc R Soc Lond B 274:1079–1086
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Taylor SE, Young JR (2006) A comparative behavioral study of three greater sage-grouse populations. Wilson J Ornithol 118:36–41
Taylor SE, Oyler-McCance SJ, Quinn TW (2003) Isolation and characterization of microsatellite loci in greater sage-grouse (Centrocercus urophasianus). Mol Ecol Notes 3:262–264
Westemeier RL, Brawn JD, Simpson SA, Esker TL, Jansen RW, Walk JW, Kershner EL, Bouzat JL, Paige KN (1998) Tracking the long-term decline and recovery of an isolated population. Science 282:1695–1698
Acknowledgments
We thank C. Overton, E. Kolada, J. Ragni, B. Prochazka, and M. Farinha, and numerous dedicated field technicians from the Western Ecological Research Center of the USGS for collecting samples for this study. Additional samples were provided by S. Espinosa of the Nevada Department of Wildlife and by S. Pellegrini and his students from Yerington High School, Yerington, Nev. Support for sample collection was also provided by Quail Unlimited; the University of Nevada, Reno; the California Department of Fish and Game; the U.S. Forest Service; and the Bureau of Land Management. We thank J. Richmond for helpful comments on this manuscript. Any use of trade, product, or firm names in this publication is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Oyler-McCance, S.J., Casazza, M.L., Fike, J.A. et al. Hierarchical spatial genetic structure in a distinct population segment of greater sage-grouse. Conserv Genet 15, 1299–1311 (2014). https://doi.org/10.1007/s10592-014-0618-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-014-0618-8