Abstract
Genes are thought to have evolved from long-lived and multiply-interactive molecules in the early stages of the origins of life. However, at that stage there were no replicators, and the distinction between interactors and replicators did not yet apply. Nevertheless, the process of evolution that proceeded from initial autocatalytic hypercycles to full organisms was a Darwinian process of selection of favourable variants. We distinguish therefore between Neo-Darwinian evolution and the related Weismannian and Central Dogma divisions, on the one hand, and the more generic category of Darwinian evolution on the other. We argue that Hull’s and Dawkins’ replicator/interactor distinction of entities is a sufficient, but not necessary, condition for Darwinian evolution to take place. We conceive the origin of genes as a separation between different types of molecules in a thermodynamic state space, and employ a notion of reproducers.






Similar content being viewed by others
Notes
For example, in some organisms the UAG codon codes for the amino acid glutamine, rather than “stop”, as in the mRNA SGC (Schultz and Yarus 1996).
Peter Godfrey-Smith’s significant book (Godfrey-Smith 2009: 63f) has a critique of replicators that is very complementary to the one offered here. Godfrey-Smith conceives of selection processes in a conceptual manifold formed by the variables “fidelity of heredity”, (H) “abundance of variation”, (V) “continuity, or smoothness of the fitness landscape”, (C) and “dependence of reproductive differences on intrinsic character” (S). Since this is a continuous space, he, too, conceives of replication as occurring at one corner of the space, rather than there being some qualitative difference between replicators and other objects in evolution.
Arguments to this effect have been made by several authors e.g., Weber (2005).
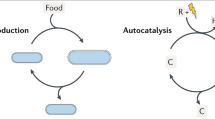
Not, as the figure might suggest (though not the caption) developmental states of a typical lifecycle.
Hull’s notion of an interactor, discussed below, is an ecological entity with economic properties.
The chief difference between our account and Dawkins (1976), chapter 2 is that for Dawkins, any molecule that could replicate itself is a replicator, but he seemed to suggest that other molecules were not “part” of the replicator. In the account given in this paper, molecules evolve stability through their involvement in interactions of differential rates and stability, and the entire process is protometabolic cf. Deamer and Weber (2010).
What counts as reproduction is open for debate. We think that it is broadly when the degree of structural similarity of progeny with parents exceeds similarity by chance, but this is not sufficient. A reproducer is any system that produces systems that are better than chance in their identity conditions (sequence identity, topological relations, etc.) but similarity is a vague notion in this context, and so the critical issue is what kind of identities matter. Reproducers are a class of entities, formed by populations of objects, whose interaction networks result in the creation and growth in number of the objects and thus the size and possibly number of the entities. Those entities with extremely high levels of fidelity can be described as interactor/replicator systems; those with merely enough reproductive fidelity to hold themselves together (through homo- and hetero-dimerisation with like molecules) are not interactor/replicator systems. Note that whether the entities are bounded (e.g., by a cell membrane) will determine whether they increase in number (cells) or just get bigger (hypercycles in the chemoton [see below]).
Kitcher outlines the explanatory schema for neo-darwinian selection in terms of genetic trajectories, initial distributions and frequencies of alleles, and fitness of alleles in a given environment and simple individual selection. The small capitals denote a core doctrine of the various Darwinian views of evolution. The overall argument is a straight nomological-deductive explanation schema in his view, as it is in Mary Williams’ and Hull’s generalised axiomatic views of evolution by selection. Kitcher also considers the views of Darwin himself as setting up a minimal Darwinism, which do not adhere to the centrality of selective mechanisms.
Unfortunately, the term “interactor” has a meaning also in chemistry, where it refers to any kind of stoichiometric or catalytic reaction between molecules. To avoid confusion, we will qualify the term where necessary.
Despite Szathmáry’s objections (Szathmáry 1988), the term “hypercycle” is here applied to any chemical cycle that is self-reproducing, and not those that are merely autocatalytic.
We are not committed to the detailed proposals of Eigen and Schuster here. Any autocatalytic process with several catalysts and reactants will exhibit the formal properties required by this argument cf. Wicken (1987).
The assumption here is that the reactor is a Malthusian reactor, i.e., one in which the rate of reaction is higher than the rate of the provision of new oligomers that act as raw material in the interactions of the hypercycle. Reactors in which the rate of reaction is less than or equal to the rate of new material are termed hyperbolic, after the growth curve. Malthusian reactors exhibit a growth curve similar to hyperbolic reactors but form a logistic curve as the raw material is exhausted, and competitive exclusion commences (i.e., as the carrying capacity is approached).
Although the efficiencies of various components will also be subjected to selection.
See Schrum et al. (2010) for a review.
David Hull, in conversation with JSW, posed this objection.
That is, if the conditions in R are neither severely Malthusian, nor entirely open to hyperbolic growth and thus free the selective coefficients.
In the context of speciation, Eldredge (1989) suggested “moremaker”, but this is a little awkward. We may quibble about some aspects of Griesemer’s definition, as Godfrey-Smith (2009) does, that the condition of “material overlap” between parent and progeny entities is too strong, and require only a causal interaction between these two entities, but that does not materially affect this paper’s argument.
Griesemer (1999) made a proposal to reduce genetics to developmental biology, which shows that reduction need not follow physical scale or containment. Reduction of theories itself is at least in part a matter of theoretical and explanatory convenience.
References
Abkevich VI, Gutin AM, Shakhnovich EI (1996) How the first biopolymers could have evolved. Proc Natl Acad Sci U S A 93(2):839–844
Alberti S (1997) The origin of the genetic code and protein synthesis. J Mol Evol 45(4):352–358
Arrhenius G, Sales B, Mojzsis S, Lee T (1997) Entropy and charge in molecular evolution-the case of phosphate. J Theor Biol 187(4):503–522
Blomberg C (1997) On the appearance of function and organisation in the origin of life. J Theor Biol 187(4):541–554
Brandon RN (1988) The levels of selection: a hierarchy of interactors. In: Plotkin H (ed) The role of behavior in evolution. MIT Press, Cambridge, pp 51–71
Brandon RN (1990) Adaptation and environment. Princeton University Press, Princeton
Chaitin GJ (1999) The unknowable, Springer series in discrete mathematics and theoretical computer science. Springer, Singapore
Cziko G (1995) Without miracles: universal selection theory and the second Darwinian Revolution. MIT Press, Cambridge
Dawkins R (1976) The selfish gene. Oxford University Press, New York
Dawkins R (1983) The extended phenotype: the long reach of the gene. Oxford University Press, Oxford
Dawkins R (1986) The blind watchmaker. Longman Scientific and Technical, Harlow
Dawkins R (1989) The selfish gene, New edn. Oxford University Press, Oxford
de Graaf RM, Visscher J, Schwartz AW (1995) A plausibly prebiotic synthesis of phosphonic acids. Nature 378(6556):474–477
Deamer D, Weber AL (2010) Bioenergetics and life’s origins. Cold Spring Harb Perspect Biol 2(2)
Dennett DC (1995) Darwin’s dangerous idea: evolution and the meanings of life. Simon and Schuster, New York
Di Giulio M (1997a) On the RNA world: evidence in favor of an early ribonucleopeptide world. J Mol Evol 45:571–578
Di Giulio M (1997b) The origin of the genetic code. Trends Biochem Sci 22(2):49–50
Dobzhansky T (1935) A critique of the species concept in biology. Philos Sci 2:344–355
Doolittle WF (1993) Sol’s world, the RNA world, our world. FASEB J 1:1–2
Eigen M (1993) The origin of genetic information: viruses as models. Gene 135(1–2):37–47
Eigen M, Schuster P (1979) The hypercycle, a principle of natural self-organization. Springer, Berlin
Eigen M, Winkler R (1981) Laws of the game: how the principles of nature govern chance, 1st American edn. Knopf, New York
Eigen M, Winkler-Oswatitsch R (1992) Steps towards life: a perspective on evolution. Oxford University Press, Oxford
Eigen M, Biebricher CK, Gebinoga M, Gardiner WC (1991) The hypercycle. Coupling of RNA and protein biosynthesis in the infection cycle of an RNA bacteriophage. Biochemistry 30(46):11005–11018
Eldredge N (1989) Macroevolutionary dynamics: species, niches, and adaptive peaks. McGraw-Hill, New York
Eldredge N (1995) Reinventing Darwin: the great evolutionary debate. Weidenfeld and Nicholson, London
Elzanowski, A (Anjay), Ostell J (2006) The genetic codes. National Center for Biotechnology Information (NCBI), 26 September 1996 [cited 11 January 2006]. Available from http://bioinformatics.org/JaMBW/2/3/TranslationTables.html
Ertem G, Ferris JP (1996) Synthesis of RNA oligomers on heterogeneous templates. Nature 379(6562):238–240
Fisher, RA (1930) The genetical theory of natural selection. Clarendon Press, Oxford (rev. ed. Dover, New York, 1958)
Ganti T (1997) Biogenesis itself. J Theor Biol 187(4):583–593
Ghiselin MT (1974) The economy of nature and the evolution of sex. University of California Press, Berkeley
Godfrey-Smith P (2009) Darwinian populations and natural selection. Oxford University Press, Oxford
Griesemer JR (1999) Materials for the study of evolutionary transition. Biol Philos 14(1):127–142
Griesemer JR (2000) The units of evolutionary transition. Selection 1(1–3):67–80
Griesemer JR (2005) The informational gene and the substantial body: on the generalization of evolutionary theory by abstraction. In: Jones MR, Cartwright N (eds) Idealization XII: correcting the model. Idealization and abstraction in the sciences. Rodopi Publishers, Amsterdam, 59–115
Griffiths PE, Gray RD (1994) Replicators and vehicles–or developmental systems. Behav Brain Sci 17(4):623–624
Griffiths PE, Gray RD (1997) Replicator II—judgement day. Biol Philos 12(4):471–492
Griffiths PE, Neumann-Held E (1999) The many faces of the gene. Bioscience 49(8):656–662
Huber C, Wächtershäuser G (1997) Activated acetic acid by carbon fixation on (Fe, Ni)S under primordial conditions. Science 276(5310):245–247
Hull DL (1974) Philosophy of biological science. Prentice-Hall, Englewood Cliffs
Hull DL (1981) Units of evolution: a metaphysical essay. In: Jensen UL, Harré R (eds) The philosophy of evolution. Harvester Press, Brighton, pp 23–44
Hull DL (1988a) Interactors versus vehicles. In: Plotkin HC (ed) The role of behavior in evolution. MIT Press, Cambridge
Hull DL (1988b) Science as a process: an evolutionary account of the social and conceptual development of science. University of Chicago Press, Chicago
Hull DL (1989) The metaphysics of evolution. State University of New York Press, Albany
Hull DL (1992) Individual. In: Keller E, Lloyd E (eds) Keywords in evolutionary biology. Harvard University Press, Cambridge, pp 180–187
Hull DL, Wilkins JS (2005) Replication. Stanford Encyclopedia of Philosophy, http://plato.stanford.edu/entries/replication/
Jablonka E, Lamb MJ (1995) Epigenetic inheritance and evolution: the Lamarckian dimension. Oxford University Press, Oxford
Jablonka E, Lamb MJ (2005) Evolution in four dimensions: genetic, epigenetic, behavioral, and symbolic variation in the history of life, Life and mind. MIT Press, Cambridge
Kauffman SA (1993) The origins of order: self-organization and selection in evolution. Oxford University Press, New York
Kauffman SA (1995) At home in the universe: the search for laws of self-organization and complexity. Oxford University Press, New York
Kauffman SA (2001) Prolegomenon to a general biology. Ann N Y Acad Sci 935:18–36; discussion 37-18
Kim J (1993) Supervenience and mind: selected philosophical essays, Cambridge studies in philosophy. Cambridge University Press, New York
Kitcher P (1993) The advancement of science: science without legend, objectivity without illusions. Oxford, New York
Lee DH, Severin K, Yokobayashi Y, Ghadiri MR (1997) Emergence of symbiosis in peptide self-replication through a hypercyclic network. Nature 390(6660):591–594
Levy M, Miller SL (1998) The stability of the RNA bases: implications for the origin of life. Proc Natl Acad Sci USA 95(14):7933–7938
Lewontin RC (1974) The genetic basis of evolutionary change, Columbia biological series no. 25. Columbia University Press, New York
Lifson S (1997) On the crucial stages in the origin of animate matter. J Mol Evol 44(1):1–8
Maclaurin J (1998) Reinventing molecular Weismannism: information in evolution. Biol Philos 13(1):37–59
Mayr E (1942) Systematics and the origin of species from the viewpoint of a zoologist. Columbia University Press, New York
Muller AW (1995) Were the first organisms heat engines? A new model for biogenesis and the early evolution of biological energy conversion. Progr Biophys Mol Biol 63(2):193–231
Muller AW (1996) Hypothesis: the thermosynthesis model for the origin of life and the emergence of regulation by Ca2+. Essays Biochem 31:103–119
Nelson KE, Levy M, Miller SL (2000) Peptide nucleic acids rather than RNA may have been the first genetic molecule. PNAS 97(8):3868–3871
Nielsen PE (1993) Peptide nucleic acid (PNA): a model structure for the primordial genetic material? Orig Life Evol Biosph V23(5):323–327
Nowak MA, Ohtsuki H (2008) Prevolutionary dynamics and the origin of evolution. Proc Natl Acad Sci 105(39):14924–14927
Oyama S (1985) The ontogeny of information: developmental systems and evolution. Cambridge University Press, Cambridge
Plotkin HC (Henry C.) (1994) Darwin machines and the nature of knowledge. In: Plotkin H. Harvard University Press, Cambridge
Poole AM, Jeffares DC, Penny D (1998) The path from the RNA world. J Mol Evol 46:1–17
Rosenberg A (1994) Instrumental biology, or, the disunity of science. University of Chicago Press, Chicago
Schneider ED, Kay JJ (1994) Life as a manifestation of the Second Law of Thermodynamics. Math Comput Model 19(6–8):25–48
Schneider ED, Sagan D (2005) Into the cool: energy flow, thermodynamics, and life. University of Chicago Press, Chicago
Schoning KU, Scholz P, Guntha S, Wu X, Krishnamurthy R, Eschenmoser A (2000) Chemical etiology of nucleic acid structure: the alpha-Threofuranosyl-(3′ → 2′) Oligonucleotide system. Science 290(5495):1347–1351
Schrum JP, Zhu TF, Szostak JW (2010) The origins of cellular life. Cold Spring Harbor Perspect Biol
Schultz DW, Yarus M (1996) On malleability in the genetic code. J Mol Evol 42:597–601
Smith JM (1975) The theory of evolution, 3rd edn. Penguin, Harmondsworth
Smith JM (2000) The concept of information in biology. Philos Sci 67(2):177–194
Smith JM, Szathmáry E (1995) The major transitions in evolution. WH Freeman/Spektrum, Oxford
Stegmann UE (2004) The Arbitrariness of the genetic code. Biol Philos 19(2):205–222
Szathmáry E (1988) A hypercyclic illusion. J Theor Biol 134(4):561–563
Szathmáry E (1997) Origins of life. The first two billion years. Nature 387(6634):662–663
Szathmáry E, Demeter L (1987) Group selection of early replicators and the origin of life. J Theor Biol 128(4):463–486
Szathmáry E, Smith JM (1997) From replicators to reproducers: the first major transitions leading to life. J Theor Biol 187(4):555–571
van Gelder T (1998) The dynamical hypothesis in cognitive science. Behav Brain Sci 21(5):616–665
Varetto L (1998) Studying artificial life with a molecular automaton. J Theor Biol 193(2):257–285
Wächtershäuser G (1997) The origin of life and its methodological challenge. J Theor Biol 187(4):483–494
Waters K (2000) Molecules made biological. Revue Internationale de Philosophie 54(4):539–564
Weber M (2005) Philosophy of experimental biology, Cambridge studies in philosophy and biology. Cambridge University Press, Cambridge
Wicken JS (1985) Thermodynamics and the conceptual structure of evolutionary theory. J Theor Biol 117(3):363–383
Wicken JS (1987) Evolution, thermodynamics, and evolution: extending the Darwinian Program. Oxford University Press, New York
Wilkins JS (2001) The appearance of Lamarckism in the evolution of culture. In: Laurent J, Nightingale J (eds) Darwinism and evolutionary economics. Edward Elgar, Cheltenham, pp 160–183
Wilkins JS (2007) The concept and causes of microbial species. Stud Hist Philos Life Sci 28(3):389–408
Williams GC (1966) Adaptation and natural selection: a critique of some current evolutionary thought. Princeton University Press, Princeton
Williams MB (1970) Deducing the consequences of evolution: a mathematical model. J Theor Biol 29:343–385
Williams GC (1992) Natural selection: domains, levels, and challenges. Oxford University Press, New York
Woese C (1998) The universal ancestor. Proc Natl Acad Sci U S A 95(12):6854–6859
Yockey HP (1992) Information theory and molecular biology. Cambridge University Press, Cambridge
Yockey HP (1995) Comments on “let there be life; thermodynamic reflections on biogenesis and evolution” by Avshalom C. Elitzur. J Theor Biol 176(3):349–355
Acknowledgments
We are grateful to the late and much-missed David Hull, Paul Griffiths, and Jim Greisemer for correspondence and conversation with JSW on this subject. Thanks also to the reviewer and to the editor, Kim Sterelny, for helpful comments.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wilkins, J.S., Stanyon, C. & Musgrave, I. Selection without replicators: the origin of genes, and the replicator/interactor distinction in etiobiology. Biol Philos 27, 215–239 (2012). https://doi.org/10.1007/s10539-011-9298-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10539-011-9298-7