Abstract
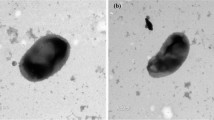
A novel bacterium, strain 1ZS3-15T, was isolated from rhizosphere of rice. Its taxonomic position was investigated using a polyphasic approach. The novel strain was observed to be Gram-stain positive, spore-forming, aerobic, motile and rod-shaped. Phylogenetic analysis based on 16S rRNA gene sequences showed that strain 1ZS3-15T was recovered within the genus Paenibacillus. It is closely related to Paenibacillus pectinilyticus KCTC 13222T (97.9 % similarity), Paenibacillus frigoriresistens CCTCC AB 2011150T (96.8 %), Paenibacillus alginolyticus JCM 9068T (96.4 %) and Paenibacillus chondroitinus DSM 5051T (95.5 %). The fatty acid profile of strain 1ZS3-15T, which showed a predominance of anteiso-C15:0 and iso-C16:0, supported the allocation of the strain into the genus Paenibacillus. The predominant menaquinone was found to be MK-7. The polar lipids profile of strain 1ZS3-15T was found to consist of diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, one unidentified lipid and two unidentified aminophospholipids. The cell wall peptidoglycan contains meso-diaminopimelic acid. Based on draft genome sequences, the DNA–DNA relatedness between strain 1ZS3-15T and the closely related species P. pectinilyticus KCTC 13222T are 24.2 ± 1.0 %, and the Average Nucleotide Identity values between the strains are 78.9 ± 0.1 %, which demonstrated that this isolate represents a new species in the genus Paenibacillus. The DNA G+C content was determined to be 45.3 mol%, which is within the range reported for Paenibacillus species. Characterisation by genotypic, chemotaxonomic and phenotypic analysis indicated that strain 1ZS3-15T represents a novel species of the genus Paenibacillus, for which the name Paenibacillus oryzisoli sp. nov. is proposed. The type strain is 1ZS3-15T (= ACCC 19783T = JCM 30487T).

Similar content being viewed by others
References
Ash C, Priest FG, Collins MD (1993) Molecular identification of RNA group 3 bacilli (ash, farrow, wallbanks and collins) using a PCR probe test. Antonie Van Leeuwenhoek 64:253–260
Cho SJ, Cho SH, Kim TS, Park SH, Kim SB, Lee GH (2015) Paenibacillus insulae sp. nov. isolated from soil. J Microbiol 53:588–591
Dong XZ, Cai MY (2001) Determination of biochemical properties. In: Dong XZ, Cai MY (eds) Manual for the systematic identification of general bacteria. Science Press, Beijing, pp 370–398 (In Chinese)
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Gao M, Xie LQ, Wang YX, Chen J, Xu J, Zhang XX, Sui XH, Gao JL, Sun JG (2012) Paenibacillus beijingensis sp. nov. a novel nitrogen-fixing species isolated from jujube garden soil. Antonie Van Leeuwenhoek 102:689–694
Guan NG, Xun Z, Ran Z, Xin YC, Zhi LC, Xue DL (2015) Paenibacillus herberti sp. nov. an endophyte isolated from herbertus sendtneri. Antonie Van Leeuwenhoek 108:587–596
Guo X, Zhou S, Wang YW, Wang HM, Kong DL, Zhu J (2016) Paenibacillus salinicaeni sp. nov. isolated from saline silt sample. Antonie Van Leeuwenhoek 109:721–728
Han TY, Tong XM, Wang YW, Wang HM, Chen XR, Kong DL (2015) Paenibacillus populi, sp. nov. a novel bacterium isolated from the rhizosphere of populus alba. Antonie Van Leeuwenhoek 108:659–666
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kim HS, Srinivasan S, Lee SS (2015) Paenibacillus alba sp. nov. isolated from peat soil. Curr Microbiol 70:865–870
Komagata K, Suzuki KI (1988) 4 Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–175
Lechevalier MP, Lechevalier HA (1980) The chemotaxonomy of actinomycetes. In: Dietz A, Thayer DW (eds) Actinomycete taxonomy (Special Publication no.6). Society for Industrial Microbiology, Arlington, pp 227–291
Lee M, Ten LN, Baek SH, Im WT, Aslam Z, Lee ST (2007) Paenibacillus ginsengisoli sp. nov. a novel bacterium isolated from soil of a ginseng field in Pocheon province, South Korea. Antonie Van Leeuwenhoek 91:127–135
Logan NA, Berge O, Bishop AH, Busse HJ, De Vos P, Fritze D, Heyndrickx M, Kämpfer P, Rabinovitch L, Salkinoja-Salonen MS, Seldin L, Ventosa A (2009) Proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria. Int J Syst Evol Microbiol 59(Pt 8):2114–2121
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2012) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:1–14
Ming H, Nie GX, Jiang HC, Yu TT, Zhou EM, Feng HG (2012) Paenibacillus frigoriresistens sp. nov. a novel psychrotroph isolated from a peat bog in heilongjiang, Northern China. Antonie Van Leeuwenhoek 102:297–305
Minnikin DE, Odonnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. Methods Microbiol 2:233–241
Park DS, Jeong WJ, Lee KH, Oh HW, Kim BC, Bae KS (2009) Paenibacillus pectinilyticus sp. nov. isolated from the gut of diestrammena apicalis. Int J Syst Evol Microbiol 59:1342–1347
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131
Ruan Z, Wang Y, Song J, Jiang S, Wang H, Li Y, Zhao B, Jiang R, Zhao B (2014) Kurthia huakuii sp. nov., isolated from biogas slurry, and emended description of the genus Kurthia. Int J Syst Evol Microbiol 64:518–521
Rzhetsky A, Nei M (1992) A simple method for estimating and testing minimum evolution trees. Mol Biol Evol 9:945–967
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101. MIDI, Newark
Schleifer KH (1985) Analysis of the chemical composition and primary structure of murein. Methods Microbiol 18:123–156
Tamaoka J, Katayama-Fujimura Y, Kuraishi H (1983) Analysis of bacterial menaqui-none mixtures by high performance liquid chromatography. J Appl Bacteriol 300:31–36
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W:improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Yang L, Ran Z, Wang R, Su Y, Lei Z, Xin Z (2016) Paenibacillus chinensis sp. nov. isolated from maize (Zea mays, L.) seeds. Antonie Van Leeuwenhoek 109:207–213
Acknowledgments
This work was supported by the Grants from National Natural Science Foundation of China (41271273), the Chinese Ministry of Science and Technology (2013AA102802) and the Key ‘‘948’’ Project of Ministry of Agriculture of China (2011-G25).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, J., Ma, XT., Gao, JS. et al. Paenibacillus oryzisoli sp. nov., isolated from the rhizosphere of rice. Antonie van Leeuwenhoek 110, 69–75 (2017). https://doi.org/10.1007/s10482-016-0777-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-016-0777-3