Abstract
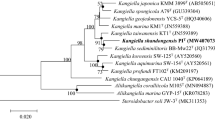
A Gram-negative bacterium, designated CAU 1040T, which was isolated from marine sand obtained from Jeju Island in South Korea, was characterized as an aerobic rod-shaped organism that that was non-motile, non-spore-forming and halophilic. The bacterium grew optimally at 37 °C, at pH 8, and in the presence of 2 % (w/v) NaCl. The taxonomic classification of CAU 1040T was investigated using a polyphasic characterization approach. While phylogenetic analysis of the 16S rRNA gene sequence revealed that CAU 1040T belongs to the genus Kangiella, the strain exhibited only 94.4–95.4 % sequence similarity to the previously described Kangiella species. Similar to other Kangiella species, Q-8 was the predominant ubiquionone and iso-C15:0 was the major cellular fatty acid detected in strain CAU 1040T. The predominant polar lipids identified were diphosphatidylglycerol, phosphatidylglycerol, and phosphatidylethanolamine. The G+C content of the CAU 1040T genome was 45.3 mol%. The phylogenetic, physiological, biochemical and chemotaxonomic data obtained in this study indicate that strain CAU 1040T represents a novel species of the genus Kangiella, for which the name Kangiella chungangensis sp. nov. is hereby proposed. The type strain is CAU 1040T (KCTC 42299T, NBRC 110728T).

Similar content being viewed by others
References
Ahn J, Park JW, McConnell JA, Ahn YB, Häggblom MM (2011) Kangiella spongicola sp. nov., a halophilic marine bacterium isolated from the sponge Chondrilla nucula. Int J Syst Evol Microbiol 61:961–964
Bowman JP (2000) Description of Cellulophaga algicola sp. nov., isolated from the surfaces of Antarctic algae, and reclassification of Cytophaga uliginosa (ZoBell and Upham 1944) Reichenbach 1989 as Cellulophaga uliginosa comb. nov. Int J Syst Evol Microbiol 50:1861–1868
Cowan ST, Steel KJ (1965) Manual for the identification of medical bacteria. Cambridge University Press, London
Da Costa MS, Albuquerque L, Nobre M, Wait R (2011) The identification of polar lipids in prokaryotes. Methods Microbiol 38:165–181
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Felsenstein J (1989) PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164–166
Fitch WM, Margoliash E (1967) Construction of phylogenetic trees. Science 155:279–284
Gordon RE, Mihm JM (1962) Identification of Nocardia caviae (Erikson) nov. comb. Ann N Y Acad Sci 98:628–636
Jean WD, Huang SP, Chen JS, Shieh WY (2012) Kangiella taiwanensis sp. nov. and Kangiella marina sp. nov., marine bacteria isolated from shallow coastal water. Int J Syst Evol Microbiol 62:2229–2234
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HH (ed) Mammalian protein metabolism. Academic Press, New York, pp 21–132
Komagata K, Suzuki K (1987) Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–208
Lane DJ (1991) 16S/23S RNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, London, pp 115–175
Lányí B (1987) Classical and rapid identification methods for medically important bacteria. Methods Microbiol 19:1–67
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Lee SY, Park S, Oh TK, Yoon JH (2013) Kangiella sediminilitoris sp. nov., isolated from a tidal flat sediment. Int J Syst Evol Microbiol 63:1001–1006
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol 3:208–218
Minnikin DE, Patel PV, Alshamaony L, Goodfellow M (1977) Polar lipid composition in the classification of Nocardia and related bacteria. Int J Syst Bacteriol 27:104–117
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Romanenko LA, Tanaka N, Frolova GM, Mikhailov VV (2010) Kangiella japonica sp. nov., isolated from a marine environment. Int J Syst Evol Microbiol 60:2583–2586
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 607–654
Tamaoka J, Komagata K (1984) Determination of DNA base composition by reverse-phase high-performance liquid chromatography. FEMS Microbiol Lett 25:125–128
Yoon JH, Oh TK, Park YH (2004) Kangiella koreensis gen. nov., sp. nov. and Kangiella aquimarina sp. nov., isolated from a tidal flat of the Yellow Sea in Korea. Int J Syst Evol Microbiol 54:1829–1835
Yoon JH, Kang SJ, Lee SY, Lee JS, Oh TK (2012) Kangiella geojedonensis sp. nov., isolated from seawater. Int J Syst Evol Microbiol 62:511–514
Acknowledgments
This research was supported by the Chung-Ang University Research Scholarship Grant in 2015.
Conflict of interest
The authors have no conflicts of interest to declare.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kim, JH., Ward, A.C. & Kim, W. Kangiella chungangensis sp. nov. isolated from a marine sand. Antonie van Leeuwenhoek 107, 1291–1298 (2015). https://doi.org/10.1007/s10482-015-0423-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-015-0423-5