Abstract
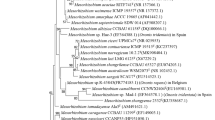
The Gram–negative, rod-shaped slow-growing strains Vaf-17, Vaf-18T and Vaf-43 were isolated from the nodules of Vavilovia formosa plants growing in the hard-to-reach mountainous region of the North Ossetian State Natural Reserve (north Caucasus, Russian Federation). The sequencing of 16S rDNA (rrs), ITS region and five housekeeping genes (atpD, dnaK, recA, gyrB and rpoB) showed that the isolated strains were most closely related to the species Bosea lathyri (class Alphaproteobacteria, family Bradyrhizobiaceae) which was described for isolates from root nodules of Lathyrus latifolius. However the sequence similarity between the isolated strains and the type strain B. lathyri LMG 26379T for the ITS region was 90 % and for the housekeeping genes it was ranged from 92 to 95 %. All phylogenetic trees, except for the rrs-dendrogram showed that the isolates from V. formosa formed well-separated clusters within the Bosea group. Differences in phenotypic properties of the B. lathyri type strain and the isolates from V. formosa were studied using the microassay system GENIII MicroPlate BioLog. Whole-cell fatty acid analysis showed that the strains Vaf-17, Vaf-18T and Vaf-43 had notable amounts of C16:0 (4.8–6.0 %), C16:0 3-OH (6.4–6.6 %), C16:1 ω5c (8.8–9.0 %), C17:0 cyclo (13.5–13.9 %), C18:1 ω7c (43.4–45.4 %), C19:0 cyclo ω8c (10.5–12.6 %) and Summed Feature (SF) 3 (6.4–8.0 %). The DNA–DNA relatedness between the strains Vaf-18T and B. lathyri LMG 26379T was 24.0 %. On the basis of genotypic and phenotypic analysis a new species Bosea vaviloviae sp. nov. (type strain RCAM 02129T = LMG 28367T = Vaf-18T) is proposed.


Similar content being viewed by others
References
Alefeld F (1866) Landwirtschaftliche Flora. Verlag Wiegand und Hempel, Berlin, p 360
Chang YL, Wang JY, Wang ET, Liu HC, Sui XH, Chen WX (2011) Bradyrhizobium lablabi sp. nov., isolated from effective nodules of Lablab purpureus and Arachis hypogaea. IJSEM 61:2496–2502
Das SK, Mishra AK, Tindall BJ, Rainey FA, Stackebrandt E (1996) Oxidation of thiosulfate by a new bacterium, Bosea thiooxidans (strain BI-42) gen. nov., sp. nov.: analysis of phylogeny based on chemotaxonomy and 16S ribosomal DNA sequencing. Int J Syst Bacteriol 46:981–987
De Meyer SE, Willems A (2012) Multilocus sequence analysis of Bosea species and description of Bosea lupini sp. nov., Bosea lathyri sp. nov. and Bosea robiniae sp. nov., isolated from legumes. IJSEM 62:2505–2510
Fedorov AA (1939) Wild high-mountain peas of Caucasus. Trans Biol Inst Arm SSR 1:39–79 (in Russian)
Goris J, Suzuki K-I, De Vos P, Nakase T, Kersters K (1998) Evaluation of a microplate DNA-DNA hybridisation method compared with the initial renaturation method. Can J Microbiol 44:1148–1153
Islam MS, Kawasaki H, Muramatsu Y, Nakagawa Y, Seki T (2008) Bradyrhizobium iriomotense sp. nov., isolated from a tumor-like root of the legume Entada koshunensis from Iriomote Island in Japan. Biosci Biotechnol Biochem 72(6):1416–1429
Kenicer G, Smýkal P, Vishyakova M, Mikić A (2009) Vavilovia formosa, an intriguing Pisum relative. Grain Legumes 51:8–12
La Scola B, Mallet M-N, Grimont PAD, Raoult D et al (2003) Bosea eneae sp. nov., Bosea massiliensis sp. nov. and Bosea vestrisii sp. nov., isolated from hospital water supplies, and emendation of the genus Bosea (Das, 1996). Int J Syst Evol Microbiol 53:15–20
Mazur A, Stasiak G, Wielbo J, Koper P, Kubik-Komar A, Skorupska A (2013) Phenotype profiling of Rhizobium leguminosarum bv. trifolii clover nodule isolates reveal their both versatile and specialized metabolic capabilities. Arch Microbiol 195(4):255–267
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Mikić A, Smýkal P, Kenicer G et al (2009) A revival of the research on beautiful vavilovia (Vavilovia formosa syn. Pisum formosum). Pisum Genet 41:34–39
Normand P, Ponsonnet C, Nesme X et al (1996) ITS analysis of prokaryotes. Mol Microbial Ecol Man 5:1–12
Novikova N, Safronova V (1992) Transconjugants of Agrobacterium radiobacter harbouring sym genes of Rhizobium galegae can form an effective symbiosis with Medicago sativa. FEMS Microbiol Lett 93:261–268
Ouattara AS, Assih EA, Thierry S, Cayol J-L, Labat M, Monroy O, Macarie H (2003) Bosea minatitlanensis sp. nov., a strictly aerobic bacterium isolated from an anaerobic digester. Int J Syst Evol Microbiol 53:1247–1251
Ponsonnet C, Nesme X (1994) Identification of Agrobacterium strains by PCR-RFLP analysis of pTi and chromosomal regions. Arch Microbiol 161:300–309
Safronova V, Tikhonovich I (2012) Automated cryobank of microorganisms: unique possibilities for long-term authorized depositing of commercial microbial strains. In: Mendez-Vilas A (ed) Microbes in applied research: current advances and challenges. World Scientific Publishing Co., Singapore, pp 331–334. doi:10.1142/9789814405041_0066
Safronova VI, Kimeklis AK, Chizhevskaya EP, Belimov AA, Andronov EE, Pinaev AG, Pukhaev AR, Popov KP, Tikhonovich IA (2014) Genetic diversity of rhizobia isolated from nodules of the relic species Vavilovia formosa (Stev.) Fed. Antonie Van Leeuwenhoek 105:389–399
Sinjushin AA, Demidenko NV, Gostimskii SA (2009) Preliminary report on taxonomical position of Vavilovia formosa (Stev.) Fed. evidenced from morphological and molecular data. Pisum Genet 41:15–20
Sneath PHA, Sokal RB (1973) Numerical taxonomy. The principles and practices of numerical classification. Freeman WH and Co., San Francisco
Steven C (1812) Orobus formosus Steven. Mem Soc Imp Nat Moscou 4:50 (In French)
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using Maximum Likelihood, evolutionary distance, and Maximum Parismony methods. Mol Biol Evol 28:2731–2739
Vincent JM (1970) A manual for the practical study of root nodule bacteria. IBP Handbook. Blackwell Scientific Publications, Oxford, pp 73–97
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173(2):697–703
Wolde-meskel E, Terefework Z, Lindström K, Frostegard A (2004) Metabolic and genomic diversity of rhizobia isolated from field standing native and exotic woody legumes in southern Ethiopia. Syst Appl Microbiol 27(5):603–611
Acknowledgments
We thank Pia Clercx for performing DNA–DNA hybridizations and excellent technical assistance. This work was partially supported by the Ministry of Education and Sciences of the Russian Federation (Agreement No. 14.604.21.0024, RFMEFI60414X0024) and by the European Union’s Seventh Framework Programme (BRIO project, grant KBBE 2010-4-266106). ITS sequencing analysis was supported by the Russian Science Foundation (Agreement No. 14-26-00094).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10482_2015_383_MOESM1_ESM.docx
Fig. S1 Phylogenetic tree of the family Bradyrhizobiaceae generated by the Neighbour-Joining method using the atpD gene sequences. Bootstrap values more than 30 % are given. Strains Bosea vaviloviae sp. nov. in bold. Type species are indicated by the letter “T” (DOCX 31 kb)
10482_2015_383_MOESM2_ESM.docx
Fig. S2 Phylogenetic tree of the family Bradyrhizobiaceae generated by the Neighbour-Joining method using the dnaK gene sequences. Bootstrap values more than 30 % are given. Strains Bosea vaviloviae sp. nov. in bold. Type species are indicated by the letter “T” (DOCX 31 kb)
10482_2015_383_MOESM3_ESM.docx
Fig. S3 Phylogenetic tree of the family Bradyrhizobiaceae generated by the Neighbour-Joining method using the recA gene sequences. Bootstrap values more than 30 % are given. Strains Bosea vaviloviae sp. nov. in bold. Type species are indicated by the letter “T” (DOCX 32 kb)
10482_2015_383_MOESM4_ESM.docx
Fig. S4 Phylogenetic tree of the family Bradyrhizobiaceae generated by the Neighbour-Joining method using the gyrB gene sequences. Bootstrap values more than 30 % are given. Strains Bosea vaviloviae sp. nov. in bold. Type species are indicated by the letter “T” (DOCX 30 kb)
10482_2015_383_MOESM5_ESM.docx
Fig. S5 Phylogenetic tree of the family Bradyrhizobiaceae generated by the Neighbour-Joining method using the rpoB gene sequences. Bootstrap values more than 30 % are given. Strains Bosea vaviloviae sp. nov. in bold. Type species are indicated by the letter “T” (DOCX 31 kb)
10482_2015_383_MOESM6_ESM.docx
Fig. S6 The unweighted pair group average clustering dendrogram showing the difference in phenotypic characteristics between the isolates from V. formosa and the type strain Bosea lathyri LMG 26379T (DOCX 56 kb)
Rights and permissions
About this article
Cite this article
Safronova, V.I., Kuznetsova, I.G., Sazanova, A.L. et al. Bosea vaviloviae sp. nov., a new species of slow-growing rhizobia isolated from nodules of the relict species Vavilovia formosa (Stev.) Fed.. Antonie van Leeuwenhoek 107, 911–920 (2015). https://doi.org/10.1007/s10482-015-0383-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-015-0383-9