Abstract
Objectives
Reproducibility of myocardial contour determination in cardiac magnetic resonance imaging is important, especially when determining T2* values per myocardial segment as a prognostic factor of heart failure or thalassemia. A method creating a composite image with contrasts optimized for drawing myocardial contours is introduced and compared with the standard method on a single image.
Materials and methods
A total of 36 short-axis slices from bright-blood multigradient echo (MGE) T2* scans of 21 patients were acquired at eight echo times. Four observers drew free-hand myocardial contours on one manually selected T2* image (method 1) and on one image composed by blending three images acquired at TEs providing optimum contrast-to-noise ratio between the myocardium and its surrounding regions (method 2).
Results
Myocardial contouring by method 2 met higher interobserver reproducibility than method 1 (P < 0.001) with smaller Coefficient of variance (CoV) of T2* values in the presence of myocardial iron accumulation (9.79 vs. 15.91 %) and in both global myocardial and mid-ventricular septum regions (12.29 vs. 16.88 and 5.76 vs. 8.16 %, respectively).
Conclusion
The use of contrast-optimized composite images in MGE data analysis improves reproducibility of myocardial contour determination, leading to increased consistency in the calculated T2* values enhancing the diagnostic impact of this measure of iron overload.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Myocardial T2* imaging has been adopted in clinical studies for non-invasive longitudinal assessment of myocardial iron deposition [1, 2]. Inferring iron deposition from magnetic resonance T2-weighted imaging requires the fitting of signal intensities from a region of interest (ROI) propagated through a series of multi-gradient echo (MGE) acquisitions to a T2* equation.
In myocardial T2* assessment, a single short-axis image that corresponds to a single echo time (TE) of a MGE series is generally selected to draw left ventricular (LV) epicardial and endocardial contours. Commercial software packages available for T2* calculation also rely on single image selection. Examples are the use of the first or second TE of the MGE image series [3] or a TE showing good contrast between the left ventricle blood pool (LVBP) and the myocardium [4].
The clarity of LV epicardial and endocardial contours depends on the contrast difference between the myocardium and LVBP defining its inner border and between the myocardium with right ventricle blood pool (RVBP), epicardial fat, abdominal fat, stomach, and lung defining its outer border [5, 6]. Reflecting the differences in transverse magnetization, a short-axis image acquired at short TE tends to have little contrast between LV myocardium and LVBP or RVBP, while at long TE it tends to show poor contrast between LV myocardium and lung. Images with long TE are subject to signal loss at regions adjacent to the posterior vein of the LV [7]. A single short-axis image cannot, therefore, provide optimal contrast between LV myocardium and all of its surroundings. Inaccurate, poorly reproducible myocardium contours are expected to result in unreliable T2* determination and, therefore, unreliable iron content representation [8].
In clinical practice, the T2* technique as a measure of myocardial iron accumulation is applied on ROIs covering either mid-ventricular septum [1] or the entire left ventricular wall on three short axis slices [2]. T2* assessment limited to the mid-ventricular septum rather than the global contour is meant to avoid susceptibility artifacts arising from the anterior and posterior cardiac vessels veins and the lung [1]. However, Meloni et al. [3, 9] showed that blood oxygenation does not significantly affect the global heart T2* values. Moreover, in a large study global heart T2* identified four groups of patients having different stages of myocardial iron loading in which the mid-ventricular septum had T2* values not matching those of the global heart [2].
The standard method for T2* assessment is manually drawing the myocardial contour on one visually selected MGE image. It is likely that contour reproducibility is affected by the choice of which MGE image is used for the tracing. As an alternative, we introduce a contrast-optimized composite image derived from those short-axis MGE images providing maximum contrast-to-noise ratio between LV myocardium and its main surrounding areas (left ventricular blood pool, right ventricular blood pool, and lung) as a new standard image in T2* assessment. The interobserver reproducibility of myocardial contouring in the T2* assessment by this new method is tested by comparing the outcomes to those made by the established manual standard method for global ROI contours as well as those limited to the mid-ventricular septum. Therefore, the purpose of this study is to provide a method for improving reproducibility of myocardial contour and T2* determination in MGE bright blood magnetic resonance imaging.
Materials and methods
Study population
This study was approved by the hospital review board, which waived informed consent since the study was retrospective and involved postprocessing of clinical data. Between February 2009 and May 2011. Twenty-one consecutive patients were examined by MGE cardiac magnetic resonance imaging in a clinical routine, including nine haematology patients (4 male, mean age 36 ± 22 year) for whom determination of T2* as a measure of myocardial iron content was requested, and twelve patients suspected of cardiomyopathy (8 male, mean age 45 ± 18 year). No patients were excluded.
Magnetic resonance
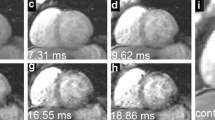
Patients were examined by cardiac magnetic resonance at a 1.5 T whole body scanner (Siemens Avanto, Erlangen, Germany) using a standard body matrix radio frequency (RF) coil of six elements and a spine matrix coil of 6–24 elements depending on the chosen field of view. A bright blood MGE sequence with a single breath hold was performed at 8 TE (2.59–18.20, and 2.23 ms increment), repetition time 200 ms, flip angle 20°, variable field of view (237–400) × 400 mm2 depending on patient size, reconstructed voxel size 1.56 × 1.56 × 10 mm3, 50 % phase resolution sampling, phase encode per cycle 5, or 16–26 cardiac cycles per breath hold, pixel bandwidth 814 Hz, and without enabling parallel imaging. In the analyses described in "Reproducibility evaluation of myocardial contouring", raw image data were used. Electrocardiogram (ECG) triggering, breath-holding together with an acquisition window shorter than the diastole duration were set to prevent motion blurring. From nine haematology patients, a total of twenty-three LV short-axis slices were acquired at basal, mid-ventricular, and apical positions of seven patients and at two mid-ventricular slices of two patients. From twelve suspected cardiomyopathy patients, eleven short-axis at mid-ventricular from eleven patients and two at mid-ventricular and basal from one patient were acquired, adding up to 36 LV short-axis slices in total. A typical dataset for mid-ventricular short-axis slice is shown in Fig. 1.
Multigradient echo series of two mid-ventricular short-axis myocardia with the lowest calculated segment T2* value of 8.20 ms represents iron loading (series a) and 24.27 ms indicates lack of iron (series b). Visual image selection (method 1; blind, independent selection by each observer) meant selection of the image where the LV myocardial wall was clearest; in series a, all four observers chose TE at 4.8 2 ms while in series b, two observers chose TE at 4.82 and 9.28 ms while the other two chose TE at 15.97 ms
Composite image formation
A custom-written software for myocardial T2* assessment was developed in MATLAB version 7.14 (The MathWorks, Natick, MA, USA). The software has the option to assess the T2* by manually drawing epicardial and endocardial contours on one subjectively selected TE image of MGE image series (Fig. 1) and on a contrast-optimized composite image.
The contrast-optimized composite images were generated first by manual drawing, on the first TE of the MGE image series, a contour of epicardial left ventricular myocardium, LVBP, RVBP, and lung with avoiding blood vessel, and on air background staying clear of any visible artefact (Fig. 2a, b). The ROI between the epicardial LV and LVBP was determined as the region of the myocardium. All contours were automatically propagated through the MGE image series and the signal-to-noise ratio (SNR) of myocardium was calculated using the following equation [10]:
where NF, SI, σ b represent the noise factor, signal intensities of the full myocardial contour representing the signal intensity of myocardium at every TE and standard deviation of air background, respectively. The noise factor in this equation accounts for the underestimation of noise derived from magnitude data and varies between 0.655 for a single RF coil and 0.71 for up to 32 coil elements [11]. Next, the contrast-to-noise ratios (CNRs) in each MGE image were calculated by using the following equation [10]:
where NF, SI s , SI m , and σ b represent the noise factor, signal intensities of the surroundings boundary of LV myocardium (LVBP [inner], RVBP [septal], and lung [anterolateral]), LV myocardium, and standard deviation of air background, respectively.
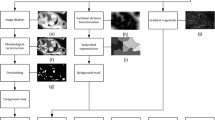
Composite image generation (method 2) as shown by two patients with (a, c, e, g), and without iron loading (b, d, f, h). First, the regions of interest (ROI) are drawn manually on the epicardial left ventricle myocardium (green), left ventricle blood pool (red), right ventricle blood pool (blue), lung (yellow), and air background (white) (a, b). Then the contrast-to-noise ratio (CNR) of the ROIs of the multi gradient echo (MGE) image series are calculated and plotted (red: LV myocardium-LVBP, blue: LV myocardium-RVBP, yellow: LV myocardium-lung) with optimal CNR values and the corresponding echo times are highlighted by circles (c, d). Three MGE images with optimal CNR, as a representation of α, β, γ, are then composed according to Eq. 3 (e, f). The composite image corresponding to C in Eq. 3 gains contrast and better demarcation, especially at the free wall of the myocardium and reduces the appearance of gradient echo susceptibility artefact at the inferior demarcation as highlighted by arrow heads (g, h)
Images with the highest mean CNR between LV myocardium with LVBP, RVBP, and lung in the MGE image series (Fig. 2c, d) were defined as optimum CNR images for composite image formation (Fig. 2e, f). The composite image (Fig. 2g, h) results from blending the registered optimum CNR images at equal weighting using the following equation:
where C corresponds to the composite image, and α, β, γ are the respective short-axis MGE images providing optimum CNR between LV myocardium and LVBP, RVBP, and lung.
T2* quantification
Pixel-wise myocardial T2* was calculated for 4–6 segments per slice according to the American Heart Association (AHA) 16-segment model [2] using a monoexponential model with offset correction [12]:
where y, K, TE, T2* and C represent signal intensity, a fitting constant, echo time, myocardium transverse relaxation time including the effect of field inhomogeneity, and a constant of offset correction (assumed to be zero, see below), respectively. For the segmental T2* analysis purposes, the AHA reference point for each slice was defined at the anterior-septal junction of LV and right ventricle [13] by the first author with more than 3 years cardiovascular imaging experience. In this study, the documentation of minimum segmental T2* value of short-axis images served to provide an indication of the presence of pathology where myocardial T2* ≤ 10 ms was considered as having severe myocardial iron, between 10 until 20 ms as having moderate myocardial iron, and >20 ms without iron loading [14, 15].
For noise determination, the evaluation of SNR of myocardium and an ROI of air background at every TE of all datasets was calculated using Eq. 1. Since in our data, even at the longest TE values, the SNR of the MGE images exceeded a value of 4, we can exclude that the T2* values calculated in monoexponential analysis were affected significantly by Rician noise [16, 17]. At SNR = 4 the correction schema proposed by Gudbjartsson et al. [18], would prescribe a small downward correction of MGE signal (−3.87 %). Therefore, Rician noise was not corrected for, and in our analyses Eq. 4 was reduced to [13]:
Reproducibility evaluation of myocardial contouring
Four observers drew short-axis LV epicardial and endocardial contours by two procedures; method 1: the clinical standard method of drawing on one visually selected MGE image with good contrast (blind, independent selection by each observer) and method 2: our novel procedure of drawing contours on the contrast-optimized composite image. Two observers had more than 3 years cardiovascular imaging experience; the other two had less experience, nonetheless they had profound knowledge of short-axis myocardium anatomy.
Hand-tracing contours were drawn independently by the observers for each of the two methods using custom-written software described above with an interval of more than 2 weeks between methods. The LV endocardial region was drawn to encompass the entire blood pool with including papillary muscles and LV trabeculae. Optimum settings of window level and width were set by the first author for method 1 and defined automatically for method 2.
In method 2, selections of MGE images for maximum CNR between myocardium and its main surroundings were observer independent. Therefore in this study, the first author used the above mentioned semi-automated procedure to generate the composite images.
Lacking a gold standard, interobserver segmentation agreement of myocardial contours by the two methods was calculated using the dice similarity coefficient (DSC), which is a simple spatial overlap index that has been used for segmentation validation and shows stronger reflection of differences in location than differences in size [19, 20]. Between two sets of binary segmentation, a minimum DSC value of 0 indicates no spatial overlap referring to no contour agreement and a maximum value of 1 referring to a complete agreement [20]. DSC was defined as follows:
where A, B represent the contour regions and ∩, + represent the intersection and addition between regions, respectively [20].
Statistical analysis
The ability of the contrast-optimized composite image in producing better CNR of myocardium and its main surroundings in one image was assessed by comparing its CNRs to the maximum CNRs between LV myocardium and LVBP, RVBP, and lung assessed on the MGE image series.
Myocardial contouring agreement between observers was assessed by the DSC and presented as medians ± median absolute deviations for all observers and observers with experience of more than 3 years and less in cardiovascular imaging. The Paired Wilcoxon test was used to compare the DSC agreement in myocardial contouring by the two methods. T2* value per segment was calculated by using Eq. 5 and presented as means ±1 SD. The interobserver reproducibility of the two methods was assessed using the Bland–Altman analysis [21] with ±1.96 SD as the limit of agreement (LoA). The two methods were compared with the same observer’s group as the DSC assessment.
On each short-axis slice, the minimum segment T2* value was identified to investigate iron deposition in patients with quantification by using method 2. Using this information, short-axis slices containing segments with T2* value <20 and >20 ms were grouped. In these slices' groups, myocardial contouring agreement and segmental T2* reproducibility of the two methods for all observers were also assessed using the Paired Wilcoxon test and Bland–Altman analysis.
The T2* quantification on the mid-ventricular septum and global heart regions was performed in this study on seven haematology patients with complete datasets of apical, mid-ventricular, and basal slices. The mid-ventricular septum was defined as the average of T2* values at mid-anterior septum and mid-inferior septum [9] and the global heart as the average of 16 segments previously described [2].
Variability between observers by using the two methods was presented as the Coefficient of variance (CoV), which is the standard deviation of the differences between observers in each method divided by their general mean and expressed as a percentage. Reliability of the methods as assessed by the observers was tested by using a two-way random intraclass correlation coefficient (ICC), evaluating absolute agreement Statistical analyses were performed using IBM SPSS Statistics software version 20 (IBM Corporation, Somers, NY, USA), P < 0.05 was considered statistically significant.
Results
T2* characteristics of patients
The minimum segmental T2* value per short-axis slice was used to assess (local) iron deposition in patients. Of the nine haematology patients, seven patients with the complete three slices had the minimum T2* in the range of 10.48–26.33 ms at apical, 8.20–27.97 ms at mid-ventricular, and 7.05–25.65 ms at basal, while two others measured at the mid-ventricular had minimum segmental T2* values of 12.72 and 17.78 ms. For the twelve suspected cardiomyopathy patients, the range of minimum segmental T2* at mid-ventricular was 12.42–36.75 ms (n = 12) and at basal 26.95 ms (n = 1).
In all 36 datasets of 21 patients, the minimum segmental T2* values <20 ms were encountered in 14 datasets of seven haematology and three suspected cardiomyopathy patients (three apical, seven mid, and four basal) in which on one haematology patient the minimum segmental T2* values <10 ms encountered in apical, mid-ventricular, and basal slices. The minimum segmental T2* values <20 ms was commonly encountered at anterior and inferior segments in apical; at anterior, inferolateral and anterolateral slices in mid-ventricular; and at anterior, anteroseptal and inferolateral in basal slices.
CNR evaluation
The contrast-optimized composite images were produced, as shown in Fig. 2, by combining those images providing maximum CNR between LV myocardium, LVBP, RVBP, and lung, generally two or three different images from the MGE series (56.71 and 41.90 % respectively). In this study, maximum CNR between LV myocardium and LVBP commonly occurred at later TE: 15.97 ms (23.40 %) and 18.20 ms (50.90 %). The pattern between LV myocardium and RVBP was similar at 15.97 ms (14.40 %) and 18.20 ms (29.90 %), while maximum CNR between LV myocardium and lung commonly happened at early TE: 2.59 ms (30.10 %) and 4.82 ms (30.80 %). Since the custom-written software only needs 1 s to generate the composite image, the total time to produce a semiautomatic composite image depends on the time needed by the observer to draw the LV epicardium, LVBP, RVBP, and lung adjacent to the myocardium (about 15 s).
In method 2, the semi-automatic analysis yielded reproducible improvements of CNR on the composite image as compared with the original MGE images. Beyond the maximum value measured at any MGE image, the composite image increased contrast by an average of 12.08 % between myocardium and LVBP (95 % CI 9.22–14.94 %; ICC of 0.89); 9.74 % between myocardium and RVBP (95 % CI 6.26–13.22 %; ICC of 0.83); and 30.33 % between myocardium and lung (95 % CI 28.21–32.45 %; ICC of 0.72). On average, between myocardium and its main surroundings, the contrast improvement was 17.38 % (95 % CI 15.64–19.13 %; ICC of 0.87) above maximum CNR at any TE.
Interobserver reproducibility of myocardial contouring
The total of 36 short-axis slices yields a set of 36 DSCs of myocardial contours of short-axis images for each method, and the combination of four observers thus adds up to sets of (36 × 6 = 216) DSCs contours. Between all observers, interobserver reproducibility of myocardial contouring improved when using contrast-optimized composite images (method 2) compared to visually selected images (method 1) with DSC of 0.844 ± 0.032 vs. 0.811 ± 0.045, P < 0.001. Between observers with more than 3 years cardiovascular imaging experience, the improvements also were significant (0.871 ± 0.017 vs. 0.829 ± 0.033, P < 0.003), as were those between observers with less experience in cardiovascular imaging (0.844 ± 0.034 vs. 0.783 ± 0.056, P < 0.003). As expected, the respective dice coefficients were slightly lower for the less experienced observers. On short-axis slices having segments with minimum T2* < 20 and >20 ms, interobserver reproducibility improved similarly when using method 2 compared to method 1 (0.842 vs. 0.822, P < 0.001 and 0.847 vs. 0.804, P < 0.001, respectively). When myocardial contouring was done by method 1, the most frequently selected MGE images were those of TE 9.28 ms (29.9 %), 7.05 ms (24.3 %), and 4.82 ms (22.9 %). In method 1, reproducibility of contours tended to be less between observers selecting a different image for ROI drawing (P < 0.001, data not shown).
Interobserver reproducibility of segmental myocardial T2*
A total of 36 short-axis slices yields two sets of 202 segments of myocardial T2* values between two observers (7 × 4 apical + 29 × 6 mid/basal) and the combination of four observers, thus, adding up to two sets of (202 × 6 = 1212) segments of myocardial T2* values for each method. Standard deviations of all T2* values listed in Table 1, are consistently smaller for method 2 than for method 1. Table 1 shows the results of Bland–Altman analysis for myocardial T2* per segment applied on all 36 slices of 21 patients by both methods. T2* quantification per segment between all observers using method 2 shows a better interobserver reproducibility with lower LoA and CoV compared to method 1 (LoA; CoV; ICC of ±6.49; 10.81 %; 0.987 vs. ±9.32; 15.25 %; 0.975). The same trend is seen between observers with more than 3 years (LoA; CoV; ICC of ±5.40; 8.63 %; 0.966 vs. ±6.53; 10.64 %; 0.954) and less experience in cardiovascular imaging (LoA; CoV; ICC of ±5.41; 9.19 %; 0.965 vs. ±10.03; 16.79 %; 0.895). For the T2* quantification per segment between all observers on 14 short-axis slices with minimum segment T2* < 20 ms (Table 1; Fig. 3a, b), method 2 reveals lower CoV than method 1 (LoA; CoV; ICC of ±4.73; 9.79 %; 0.994 vs. ±7.80; 15.91 %; 0.984) with the same trend on 22 short-axis slices with minimum segment T2* > 20 ms (Table 1; Fig. 3c, d) (LoA; CoV; ICC of ±7.38; 10.94 %; 0.973 vs. ±10.16; 14.77 %; 0.948).
Bland–Altman plots between observers assessing myocardial T2* on short-axis slices. a, b: Assessed on a visually selected multi gradient echo image (MGE) (method 1) and b contrast-optimized composite image (method 2) in the presence of a segment with minimum T2* < 20 ms (468 segments). c, d: Assessed on c visually selected MGE image and d contrast-optimized composite image (method 2) in the absence of segments with minimum T2* < 20 ms (744 segments)
In Table 2, the global heart and mid-ventricular septum T2* analysis was done on seven haematology patients who had complete basal, mid-ventricular, and apical slices by using the two methods. Method 2 has improved interobserver reproducibility for all observers compared to method 1 in assessing T2* on the global heart region (CoV; ICC of 12.29 %; 0.987 vs. 16.88 %; 0.976) as well as mid-ventricular septum region (CoV; ICC of 5.76 %; 0.998 vs. 8.16 %; 0.995). These improvements were also seen in observers with either high or low experience in cardiovascular imaging.
T2* analysis of all 16 myocardial segments as assessed by all observers on all 36 short-axis slices of the 21 patients is shown in Table 3. In this analysis, higher interobserver reproducibility of T2* per segment is shown by method 2 with lower CoV and higher ICC compared to method 1 in most of the 16 segments. When the T2* analysis of the mid-ventricular slice is extended from 7 (Table 2) to 21 slices, higher interobserver reproducibility by method 2 is retained in the mid-ventricular anteroseptal (CoV; ICC of 6.19 %; 0.995 vs. 7.87 %; 0.992) and inferoseptal (CoV; ICC of 5.66 %; 0.994 vs. 8.57 %; 0.986) segments corresponding with the mid-ventricular septum.
Discussion
This study shows on average 17 % CNR improvement using the proposed contrast-optimized composite image (method 2) over any maximum contrast provided by the single image technique (method 1), providing reproducible T2* values in short-axis slices of the mid-ventricular septum as well as global heart regions. These findings may be beneficial for the use of T2* imaging in the diagnosis of thalassemia and suspected cardiomyopathy patients with borderline myocardial T2* distribution. Iron overload may be heterogeneous with septal T2* values not necessarily representative for the whole left ventricular myocardium [2].
A recent study by House et al. [22] on the post mortem heart reveals segmental variation of iron loading other than septal, especially in the lateral wall, with different distribution on epicardial and endocardial regions confirming previous findings [23, 24]. Therefore, in this study, segmental T2* was assessed for all myocardial segments rather than a c-shaped area with two arbitrary cut-offs, which, although frequently used in previous studies [1], may be erroneous. Note that beside iron content, T2* variations between myocardial segments may also reflect the different orientations of the myocardial capillaries to the external magnetic field, resulting in locally varying susceptibility effects and consequently varying degrees of dephasing [25]. However, findings by Meloni et al. [3, 9], with use of artifact correction maps, indicate that neither additive susceptibility artifacts nor blood oxygenation significantly affects the global heart T2*.
Evaluation of the T2* quantification at mid-ventricular septum and global heart regions (Table 2) shows that method 2 improves the interobserver reproducibility (lower CoV and higher ICC) compared to method 1 showing the superiority of contrast-optimized composite images from a single TE image for clinical T2* quantification. Looking at the 16 myocardial segments (Table 3), T2* quantification by using method 2 also shows lower CoV and higher ICC compared to method 1 in most segments.
Comparing the results using method 2 to other research groups for global heart T2* analysis (Table 2), similar interobserver reproducibility was noted by one research group using a single TE image on its myocardial contouring [26]. While at mid-ventricular septum T2* analysis, higher interobserver reproducibility was achieved by using method 2 compared to other research groups, which used a single TE image [5, 26]. If the comparison is broken down into 16 segments T2* rather than global heart T2* (Table 3), the interobserver reproducibility by using method 2 has shown a higher interobserver reproducibility, compared to the selection of one MGE image by another research group [26]. Moreover, the selection of a good contrast (higher TE) image by method 1 provides, in overall, a better interobserver reproducibility compared to others using the first or second (short TE) image [3, 26]. This may reflect the fact that in this study, even at higher TE’s, the MGE images still had SNR > 4.
It was shown in this study that maximum contrast between myocardium and its surrounding borders is not likely to occur on a single MGE image. The observer’s preference in selecting an MGE image having a good contrast also reveals a range of TE values between 4.82 and 9.28 ms. The observer dependent difference in image selection did significantly contribute to the reduction of observers agreement on myocardial contouring (DSC value, P < 0.001) leading to higher variability (CoV) and lower reliability (ICC) in T2* quantification in this study (Method 1) (data not shown).
In this study, myocardial contouring was performed by the normal standard method (method 1) and by our new method (method 2). In general, both methods showed good interobserver reproducibility with good overlap of myocardial contour drawing. However, even better reproducibility was obtained by method 2(P < 0.001) especially for experienced observers getting DSC above 0.85. This demonstrates the consistency of myocardial information acquired by the method [19]. This result is in line with the reductions of T2*’s standard deviation, limits of agreement, and coefficients of variance also with the higher intra class correlation evaluates between observers using method 2 compared to method 1 (Table 1).
This study documented the minimum segment T2* values in order to provide an indication of the presence of pathology, such as an increased risk of heart failure at T2* < 20 ms [14]. In slices without any segment meeting T2* < 20 ms, i.e., without an indication of iron accumulation in the left ventricular myocardium, the DSC values by the two methods showed good contour overlaps between observers but higher DSC values were reached by using method 2 (P < 0.001). Importantly, in the remaining slices with segments of T2* < 20 ms indicating iron deposition as a result of pathology, the respective values were similar (P < 0.001). These results thus indicate that our newly proposed composite image method not only gives improvement in “normals” but also improves the reproducibility of myocardial contours in the presence of iron deposition as a result of focal pathology.
In this study we thus demonstrated that the use of a contrast-optimized composite image derived from the MGE image series, as a reference image for drawing myocardial contours, leads to higher segmental T2* value reproducibility compared to the use of one visually selected MGE image. The composite image is able to optimize the contrast between the myocardium and its surroundings (mainly LVBP, RVBP and lung tissue) on one image with even higher CNR (17 % in this study) than any individual maximum contrast between the myocardium and its surroundings, which are unlikely to be reached at one TE value. An equal weighting factor to generate contrast-optimized composite image (Eq. 3) is used in this study to maintain equal contrast contribution between the myocardium and its main surroundings provided by the MGE images. Seeing that maximum CNR between LV myocardium, LVBP, and RVBP (α and β in Eq. 3) happen with the same pattern on MGE images at the later TE while between LV myocardium and lung (γ in Eq. 3) happens mostly at the early TE, a combination with different weighting factors for α, β and γ can be a future direction to optimized contrast on the composite image. The short time needed for generating the composite image (on average 15 s) turns our proposed method into a good alternative for providing reference images for myocardial contour drawings in the clinical practice. The Bland–Altman limit of agreement (Table 1) show that the use of a contrast-optimized composite image is able to produce more accurate and reliable segmental T2* values obtained by different observers with lower CoV and higher ICC. Moreover, the use of the proposed contrast-optimized composite image also improves the ability of less experienced observers in assessing reproducible segmental T2*.
Our novel approach has the limitation that it was done on a small number of patient data for methodological purpose. A validation on a larger patient data base is encouraged to confirm the advantages of our new analysis method in clinical practice. The approach still requires manual drawing of the myocardial and its surrounding contours for composite image formation, which gives an additional task in the clinical routine workflow. Moreover, the ROIs of lung and air background must be carefully positioned to avoid any influence of blood vessels and ghosting artefacts. However, we expect that in the near future standardizing ROI drawing in an automatic manner will minimize this limitation. The MGE MRI protocol of this study did not include parallel imaging acquisition. In the latter case (as also to some extent with our use of phased array coils) uneven k-space sampling would make noise estimates tricky. However, in parallel imaging MGE studies the use of relative CNR’s rather than absolute CNR’s (Eq. 2) would be a valid alternative still allowing for wide implementation of our newly proposed method. Elsewhere, an automatic segmentation method for myocardial contouring showed highly improved reproducibility for T2* quantification for images showing high contrast between the myocardium and its surroundings as in black blood T2* acquisition [27]. An evaluation of the use of contrast-optimized composite images in black blood T2* acquisition would be needed to investigate the advantage of our method in producing more reproducible segmentation on the black blood image (beyond the scope of the current study). The development and implementation of automatic myocardial segmentation rather than manual ROI drawing on bright blood T2* MGE images, using the composite image method, are the topic of ongoing investigations at our institution.
Conclusions
In conclusion, the proposed contrast-optimized composite method has proven to be effective by providing highly reproducible myocardial contours yielding more accurate T2* measurements not only in the presence of myocardial iron accumulation but also in global heart T2* and mid-ventricular septum T2* analysis. This new method profits from a standardized generation of a composite bright blood T2* MGE image, with on average 17 % improved contrast to noise ratio between LV myocardium and the surrounding tissues. This improves reproducibility of myocardial contour determination, leading to increased consistency in the calculated T2* values enhancing the diagnostic impact of this measure of iron overload.
References
He T (2014) Cardiovascular magnetic resonance T2* for tissue iron assessment in the heart. Quant Imaging Med Surg 4:407–412
Meloni A, Restaino G, Borsellino Z, Caruso V, Spasiano A, Zuccarelli A, Valeri G, Toia P, Salvatori C, Positano V, Midiri M, Pepe A (2014) Different patterns of myocardial iron distribution by whole-heart T2* magnetic resonance as risk markers for heart complications in thalassemia major. Int J Cardiol 177:1012–1019
Meloni A, Positano V, Pepe A, Rossi G, Dell’Amico M, Salvatori C, Keilberg P, Filosa A, Sallustio G, Midiri M, D’Ascola D, Santarelli MF, Lombardi M (2010) Preferential patterns of myocardial iron overload by multislice multiecho T*2 CMR in thalassemia major patients. Magn Reson Med 64:211–219
Mavrogeni S, Bratis K, van Wijk K, Kyrou L, Kattamis A, Reiber JHC (2013) The reproducibility of cardiac and liver T2* measurement in thalassemia major using two different software packages. Int J Cardiovasc Imaging 29:1511–1516
He T, Gatehouse PD, Kirk P, Tanner MA, Smith GC, Keegan J, Mohiaddin RH, Pennell DJ, Firmin DN (2007) Black-blood T2* technique for myocardial iron measurement in thalassemia. J Magn Reson Imaging 25:1205–1209
Petitjean C, Dacher J-N (2011) A review of segmentation methods in short axis cardiac MR images. Med Image Anal 15:169–184
Reeder SB, Faranesh AZ, Boxerman JL, McVeigh ER (1998) In vivo measurement of T*2 and field inhomogeneity maps in the human heart at 1.5 T. Magn Reson Med 39:988–998
Brittenham GM, Badman DG (2003) Noninvasive measurement of iron: report of an NIDDK workshop. Blood 101:15–19
Meloni A, Pepe A, Positano V, Favilli B, Maggio A, Capra M, Lo Pinto C, Gerardi C, Santarelli MF, Midiri M, Landini L, Lombardi M (2009) Influence of myocardial fibrosis and blood oxygenation on heart T2* values in thalassemia patients. J Magn Reson Imaging 29:832–837
Firbank MJ, Coulthard A, Harrison RM, Williams ED (1999) A comparison of two methods for measuring the signal to noise ratio on MR images. Phy Med Biol 44:N261–N264
Dietrich O, Raya JG, Reeder SB, Ingrisch M, Reiser MF, Schoenberg SO (2008) Influence of multichannel combination, parallel imaging and other reconstruction techniques on MRI noise characteristics. Magn Reson Imaging 26:754–762
Ghugre NR, Enriquez CM, Coates TD, Nelson MD, Wood JC (2006) Improved R2* measurements in myocardial iron overload. J Magn Reson Imaging 23:9–16
Positano V, Pepe A, Santarelli MF, Scattini B, De Marchi D, Ramazzotti A, Forni G, Borgna-Pignatti C, Lai ME, Midiri M, Maggio A, Lombardi M, Landini L (2007) Standardized T2* map of normal human heart in vivo to correct T2* segmental artefacts. NMR Biomed 20:578–590
Wood JC (2007) Magnetic resonance imaging measurement of iron overload. Curr Opin Hematol 14:183–190
Carpenter J-P, Roughton M, Pennell DJ (2013) International survey of T2* cardiovascular magnetic resonance in β-thalassemia major. Haematologica 98:1368–1374
Baron P, Dorrius MD, Kappert P, Oudkerk M, Sijens PE (2010) Diffusion-weighted imaging of normal fibroglandular breast tissue: influence of microperfusion and fat suppression technique on the apparent diffusion coefficient. NMR Biomed 23:399–405
He T, Gatehouse PD, Kirk P, Mohiaddin RH, Pennell DJ, Firmin DN (2008) Myocardial T*2 measurement in iron-overloaded thalassemia: an ex vivo study to investigate optimal methods of quantification. Magn Reson Med 60:350–356
Gudbjartsson H, Patz S (1995) The Rician distribution of noisy MRI data. Magn Reson Med 34:910–914
Albà X, Figueras I, Ventura RM, Lekadir K, Tobon-Gomez C, Hoogendoorn C, Frangi AF (2014) Automatic cardiac LV segmentation in MRI using modified graph cuts with smoothness and interslice constraints. Magn Reson Med 72:1775–1784
Grosgeorge D, Petitjean C, Caudron J, Fares J, Dacher J-N (2011) Automatic cardiac ventricle segmentation in MR images: a validation study. Int J Comput Assist Radiol Surg 6:573–581
Bland JM, Altman DG (1986) Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1:307–310
House MJ, Fleming AJ, de Jonge MD, Paterson D, Howard DL, Carpenter J-P, Pennell DJ, St Pierre TG (2014) Mapping iron in human heart tissue with synchrotron x-ray fluorescence microscopy and cardiovascular magnetic resonance. J Cardiovasc Magn Reson 16:80
Ghugre NR, Enriquez CM, Gonzalez I, Nelson MD, Coates TD, Wood JC (2006) MRI detects myocardial iron in the human heart. Magn Reson Med 56:681–686
Olson LJ, Edwards WD, McCall JT, Ilstrup DM, Gersh BJ (1987) Cardiac iron deposition in idiopathic hemochromatosis: histologic and analytic assessment of 14 hearts from autopsy. J Am Coll Cardiol 10:1239–1243
Ziener CH, Kampf T, Reents G, Schlemmer H-P, Bauer WR (2012) Spin dephasing in a magnetic dipole field. Phy Rev E: Stat Nonlinear Soft Matter Phy 85:051908
Ramazzotti A, Pepe A, Positano V, Rossi G, De Marchi D, Brizi MG, Luciani A, Midiri M, Sallustio G, Valeri G, Caruso V, Centra M, Cianciulli P, De Sanctis V, Maggio A, Lombardi M (2009) Multicenter validation of the magnetic resonance T2* technique for segmental and global quantification of myocardial iron. J Magn Reson Imaging 30:62–68
Zheng Q, Feng Y, Wei X, Feng M, Chen W, Lu Z, Xu Y, Chen H, He T (2015) Automated interventricular septum segmentation for black-blood myocardial T2* measurement in thalassemia. J Magn Reson Imaging 41:1242–1250
Acknowledgments
Authors’contribution
Protocol/project development: Triadyaksa, Handayani, Oudkerk, Sijens. Data collection or management: Triadyaksa, Handayani, Willems, Dijkstra, Aryanto, Pelgrim, Xie. Data analysis: Triadyaksa, Handayani, Prakken, Oudkerk, Sijens.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
For this type of study, formal consent is not required.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Triadyaksa, P., Handayani, A., Dijkstra, H. et al. Contrast-optimized composite image derived from multigradient echo cardiac magnetic resonance imaging improves reproducibility of myocardial contours and T2* measurement. Magn Reson Mater Phy 29, 17–27 (2016). https://doi.org/10.1007/s10334-015-0503-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10334-015-0503-6