Abstract
Background
Early diagnosis and treatment of non-Hodgkin lymphoma (NHL) are progressively important. It has been shown that aberrant promoter methylation contributes to the development and progression of lymphoma. We tried to explore the effect of methylation of p16 and shp1 genes in plasma in the diagnosis of B-NHL patients.
Methods
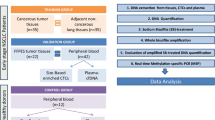
The methylation of p16 and shp1 genes in plasma were detected by methylation specific polymerase chain reaction in 103 patients with B-NHL, and compared with peripheral blood leukocytes (PBLs) and formaldehyde-fixed paraffin-embedded (FFPE) tumor tissues.
Results
The results showed that methylation frequency of p16 in plasma, PBLs, and FFPE tumor tissues of newly diagnosed B-NHL patients were 37% (27/73), 16% (12/73) and 39% (16/41), whereas those of shp1 were 47% (34/73), 25% (18/73) and 63% (26/41). High methylation consistency of p16/shp1 between plasma and FFPE tumor tissues were revealed (the values of kappa: 0.84, 0.80). Moreover, there were a higher frequency of methylated p16 in all three samples in patients with B symptoms and lower platelet count (<100 × 109/L), as well as in patients with stage III/IV in plasma and FFPE tumor tissues. Meanwhile, higher frequency of methylated shp1 was observed in patients with higher LDH level in all three samples.
Conclusion
Methylation of p16/shp1 in plasma can represent their methylation status in tumor tissues, and may be promising biomarkers in early diagnosis and prognosis evaluation in B-NHL.

Similar content being viewed by others
References
Schwarzenbach H, Hoon DS, Pantel K (2011) Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 11:426–437
Maunakea AK, Chepelev I, Zhao K (2010) Epigenome mapping in normal and disease states. Circ Res 107:327–339
Sato H, Oka T, Shinnou Y et al (2010) Multi-step aberrant CpG island hyper-methylation is associated with the progression of adult T-cell leukemia/lymphoma. Am J Pathol 176:402–415
Shi H, Guo J, Duff DJ et al (2007) Discovery of novel epigenetic markers in non-Hodgkin’s lymphoma. Carcinogenesis 28:60–70
Eberle FC, Rodriguez-Canales J, Wei L et al (2011) Methylation profiling of mediastinal gray zone lymphoma reveals a distinctive signature with elements shared by classical Hodgkin’s lymphoma and primary mediastinal large B-cell lymphoma. Haematologica 96:558–566
Amara K, Trimeche M, Ziadi S et al (2008) Prognostic significance of aberrant promoter hypermethylation of CpG islands in patients with diffuse large B-cell lymphomas. Ann Oncol 19:1774–1786
Hagiwara K, Li Y, Kinoshita T et al (2010) Aberrant DNA methylation of the p57KIP2 gene is a sensitive biomarker for detecting minimal residual disease in diffuse large B cell lymphoma. Leuk Res 34:50–54
Stewart DJ, Issa JP, Kurzrock R et al (2009) Decitabine effect on tumor global DNA methylation and other parameters in a phase I trial in refractory solid tumors and lymphomas. Clin Cancer Res 15:3881–3888
Villuendas R, Sanchez-Beato M, Koh JC et al (1998) Loss of p16/INK4A protein expression in non-Hodgkin’s lymphomas is a frequent finding associated with tumor progression. Am J Pathol 153:887–897
Oka T, Ouchida M, Koyama M (2002) Gene silencing of the tyrosine phosphatase SHP1 gene by aberrant methylation in leukemias/lymphomas. Cancer Res 62:6390–6394
Oka T, Yoshino T, Hayashi K et al (2001) Reduction of hematopoietic cell-specific tyrosine phosphatase SHP-1 gene expression in natural killer cell lymphoma and various types of lymphomas/leukemias: combination analysis with cDNA expression array and tissue microarray. Am J Pathol 159:1495–1505
Cheson BD, Pfistner B, Juweid ME et al (2007) Revised response criteria for malignant lymphoma. J Clin Oncol 25(5):579–586
El Messaoudi S, Rolet F, Mouliere F et al (2013) Circulating cell free DNA: preanalytical considerations. Clin Chim Acta 424:222–230
Krajnović M, Radojković M, Davidović R et al (2013) Prognostic significance of epigenetic inactivation of p16, p15, MGMT and DAPK genes in follicular lymphoma. Med Oncol 30:441
Krajnović M, Radojković M, Davidović R et al (2007) Quantification of circulating plasma DNA fragments as tumor markers in patients with esophageal cancer. Anticancer Res 27:2737–2742
Liggett TE, Melnikov A, Yi Q et al (2011) Distinctive DNA-methylation patterns of cell-free plasma DNA in women with malignant ovarian tumors. Gynecol Oncol 120:113–120
Sturgeon SR, Balasubramanian R, Schairer C et al (2012) Detection of promoter methylation of tumor suppressor genes in serum DNA of breast cancer cases and benign breast disease controls. Epigenetics 7:1258–1267
Ellinger J, Albers P, Perabo FG et al (2009) CpG island hypermethylation of cell-free circulating serum DNA in patients with testicular cancer. J Urol 182:324–329
Hoffmann AC, Vallböhmer D, Prenzel K et al (2009) Methylated DAPK and APC promoter DNA detection in peripheral blood is significantly associated with apparent residual tumor and outcome. J Cancer Res Clin Oncol 135:1231–1237
Lee TH, Montalvo L, Chrebtow V et al (2001) Quantitation of genomic DNA in plasma and serum samples: higher concentrations of genomic DNA found in serum than in plasma. Transfusion 41:276–282
Li L, Choi JY, Lee KM et al (2012) DNA methylation in peripheral blood: a potential biomarker for cancer molecular epidemiology. J Epidemiol 22:384–394
Teschendorff AE, Menon U, Gentry-Maharaj A et al (2009) An epigenetic signature in peripheral blood predicts active ovarian cancer. PLoS One 4:e8274
Pedersen KS, Bamlet WR, Oberg AL et al (2011) Leukocyte DNA methylation signature differentiates pancreatic cancer patients from healthy controls. PLoS One 6:e18223
Wang L, Aakre JA, Jiang R et al (2010) Methylation markers for small cell lung cancer in peripheral blood leukocyte DNA. J Thorac Oncol 5:778–785
Deligezer U, Yaman F, Erten N et al (2003) Frequent copresence of methylated DNA and fragmented nucleosomal DNA in plasma of lymphoma patients. Clin Chim Acta 335:89–94
Cosialls AM, Santidrián AF, Coll-Mulet L et al (2012) Epigenetic profile in chronic lymphocytic leukemia using methylation-specific multiplex ligation-dependent probe amplification. Epigenomics 4:491–501
Tsirigotis P, Pappa V, Labropoulos S et al (2006) Mutational and methylation analysis of the cyclin-dependent kinase 4 inhibitor (p16INK4A) gene in chronic lymphocytic leukemia. Eur J Haematol 76:230–236
Zainuddin N, Kanduri M, Berglund M et al (2011) Quantitative evaluation of p16INK4a promoter methylation using pyrosequencing in de novo diffuse large B-cell lymphoma. Leuk Res 35:438–443
Shiozawa E, Takimoto M, Makino R et al (2006) Hypermethylation of CpG islands in p16 as a prognostic factor for diffuse large B-cell lymphoma in a high-risk group. Leuk Res 30:859–867
Kim SS, Choi YH, Han CW et al (2009) DNA methylation profiles of MGMT, DAPK1, hMLH1, CDH1, SHP1, and HIC1 in B-Cell lymphomas. Korean J Pathol 43:420–427
Koyama M, Oka T, Ouchida M et al (2003) Activated proliferation of B-cell lymphomas/leukemias with the SHP1 gene silencing by aberrant CpG methylation. Lab Invest 83:1849–1858
Acknowledgements
This work was supported by grants from Tianjin cancer research programs of major projects foundation (Grant Nos. 12ZCDZSY18000), National Natural Science Foundation of China (Grant Nos. 81570106, 81570111, 81500101, 81600093, 81600088), the Tianjin Municipal Natural Science Foundation (Grant Nos. 14JCYBJC25400,15JCYBJC24300) and Tianjin Health and Family Planning Commision (Grant Nos. 15KG150).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There are no conflicts of interests.
Additional information
K. Ding and X. Chen contributed equally to this work.
About this article
Cite this article
Ding, K., Chen, X., Wang, Y. et al. Plasma DNA methylation of p16 and shp1 in patients with B cell non-Hodgkin lymphoma. Int J Clin Oncol 22, 585–592 (2017). https://doi.org/10.1007/s10147-017-1100-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10147-017-1100-7