Abstract
There is considerable interest in the use of enantioselective alcohol dehydrogenases for the production of enantio- and diastereomerically pure diols, which are important building blocks for pharmaceuticals, agrochemicals and fine chemicals. Due to the need for a stable alcohol dehydrogenase with activity at low-temperature process conditions (30°C) for the production of (2S,5S)-hexanediol, we have improved an alcohol dehydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus (AdhA). A stable S-selective alcohol dehydrogenase with increased activity at 30°C on the substrate 2,5-hexanedione was generated by laboratory evolution on the thermostable alcohol dehydrogenase AdhA. One round of error-prone PCR and screening of ∼1,500 mutants was performed. The maximum specific activity of the best performing mutant with 2,5-hexanedione at 30°C was tenfold higher compared to the activity of the wild-type enzyme. A 3D-model of AdhA revealed that this mutant has one mutation in the well-conserved NADP(H)-binding site (R11L), and a second mutation (A180V) near the catalytic and highly conserved threonine at position 183.
Similar content being viewed by others
Introduction
Alcohol dehydrogenases (ADHs) are present in all organisms. They display a wide variety of substrate specificities and play an important role in a broad range of physiological processes (Reid and Fewson 1994). There is considerable interest in stable alcohol dehydrogenases for a range of applications in food, pharmaceutical and fine chemicals industries. The production of enantio- and diastereomerically pure diols is particularly desired because these are important chemical building blocks (Haberland et al. 2002a; Hummel 1999; Radianingtyas and Wright 2003; Zeikus et al. 1998). (2S,5S)-Hexanediol is such a versatile building block for the synthesis of various fine chemicals, pharmaceuticals and chiral phosphine ligands (Haberland et al. 2002b; Brunel and Faure 2004).
For most applications robust biocatalysts are desired. Enzymes from hyperthermophiles, i.e., microorganisms that grow optimally above 80°C, generally display an extreme stability at high temperature, high pressure, as well as high concentrations of chemical denaturants (Vieille and Zeikus 2001). Enzymes are environmental friendly, biodegradable, efficient, and are of low cost in terms of resource requirements; as such they provide benefits compared to traditional chemical approaches in various industrial processes. In many instances, however, natural enzymes do not perform optimally in a particular unnatural process, and as such may not be suitable for large-scale industrial applications (Schoemaker et al. 2003; Schmid et al. 2001).
The production of (2S,5S)-hexanediol, can be achieved by chemical synthesis or biocatalytic processes. Chemical synthesis of (2,5)-hexanediol results in a racemic / meso mixture of (2,5)-hexanediol with a theoretical yield of 25% for (2S,5S)-hexanediol. This diol can also be obtained by the enzymatic reduction of the cheap 2,5-hexanedione with baker’s yeast or an enantioselective alcohol dehydrogenase (Lieser 1983). The enzymatic conversion of 2,5-hexanedione to (2S,5S)-hexanediol is preferably performed with low cost process conditions such as a process temperature of 30°C, using a stable and enantioselective alcohol dehydrogenase and an efficient cofactor regeneration system, which also allows easy purification of the end product.
Previously, the adhA gene from the hyperthermophilic archaeon Pyrococcus furiosus was cloned, sequenced and functionally overexpressed in Escherichia coli (Van der Oost et al. 2001). AdhA was purified, characterized and found to be a homodimer of 26-kDa subunits. The enzyme is very well adapted to the high growth temperature of the organism (70–103°C) (Fiala and Stetter 1986) with an optimum temperature for activity of 90°C, and a high resistance to thermal inactivation, which was shown by half-life values of 150 h at 80°C, 22.5 h at 90°C and 25 min at 100°C. AdhA is aNADP+-dependent alcohol dehydrogenase that converts alcohols to the corresponding aldehydes/ketones and vice versa, with a rather broad substrate specificity. Maximal specific activities were observed with 2-pentanol (46 U mg−1) and pyruvaldehyde (32 U mg−1) in the oxidative and reductive reaction, respectively (Van der Oost et al. 2001).
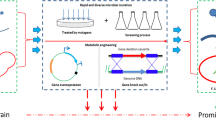
Here, we report that the activity of AdhA at 30°C does not exceed 5% of its optimal activity. In addition, AdhA has been demonstrated to be S-selective: it stereoselectively reduces 2,5-hexanedione, to (2S,5S)-hexanediol. Interestingly, it has been found that in the presence of excess 2-propanol, AdhA can perform substrate-coupled NADPH regeneration, as has been described for other ADHs (T. Dauβmann, personal communication). In substrate-coupled NADPH regeneration a single enzyme (AdhA) is involved in both the main and recycling reactions. In the oxidation of 2-propanol to acetone the coenzyme is regenerated in the same active site that carries out the reductive conversion of the substrate of interest, 2,5-hexanedione (Fig. 1). To achieve high conversions the 2-propanol should be present in excess (Bastos et al. 1999; Eckstein et al. 2004). The main goal has been the improvement of the low-temperature activity of AdhA from the hyperthermophilic archaeon Pyrococcus furiosus. There are two main routes to improve an enzyme: (1) rationale-based mutagenesis, and (2) laboratory evolution. To allow rationale-based engineering, a high-resolution crystal structure and insight into the structure-function relations of the biocatalyst of interest are required. Unfortunately, no crystal structure of AdhA is available at present. Laboratory evolution on the other hand offers a way to optimize enzymes by random mutagenesis in the absence of structural or mechanistic information (Bornscheuer and Pohl 2001; Yuan et al. 2005; Lebbink et al. 2000). The latter approach has been selected as the method to improve the low-temperature activity of AdhA on 2,5-hexanedione, while retaining its stability. The obtained improved AdhA variants have been analyzed, and the results are interpreted by using a structural model of the enzyme.
Materials and methods
Chemicals and plasmids
2,5-Hexanedione and (2S,5S)-hexanediol were kindly provided by Julich Chiral Solutions (Jülich, Germany). All other chemicals (analytical grade) were purchased from Sigma-Aldrich (Munich, Germany) or Acros Organics (Geel, Belgium).
The restriction enzymes were obtained from Invitrogen (Paisley, UK) and New England Biolabs (Ipswich, MA, USA). T4 DNA ligase and Genemorph random mutagenesis kit were purchased from Stratagene (Amsterdam, The Netherlands) and REDTaq DNA polymerase from Sigma-Aldrich. For heterologous expression the vector pET-24d (KanR; Novagen, Darmstadt, Germany) and the tRNA helper plasmid pSJS1244 (SpecR) (Kim et al. 1998; Sorensen et al. 2003) were used.
Organisms and growth conditions
Escherichia coli XL1-Blue (Stratagene) was used as a host for the construction of pET24d derivatives. E. coli BL21(DE3) (Novagen) harboring the tRNA helper plasmid pSJS1244 was used as an expression host. Both strains were grown under standard conditions (Sambrook et al. 1989) following the instructions from the manufacturer.
Construction of a random mutant AdhA library
The adhA gene (705 bp) was PCR amplified from the chromosomal DNA of P. furiosus using the primers BG1394 (sense, 5′-GCGCGCCATG G CAAAGGTTGCCGTAATTACTGGG) and BG1396 (antisense, 5′-GCGCGGGATCCTCAATACTCAGGTTTTTGATAAATTGAG), containing the NcoI and BamHI sites (underlined in the sequences). In order to introduce an NcoI restriction site an extra alanine codon (GCA) was introduced in the adhA gene by the forward primer BG1394 (boldface in the sequence). The fragment generated was purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany). The purified gene was digested with NcoI-BamHI and cloned into E. coli XL1-Blue using an NcoI-BamHI digested pET24d vector. The resulting plasmid was named pWUR104 and the sequence was confirmed by sequence analysis of both DNA strands (Baseclear, Leiden, The Netherlands).
Random mutations were introduced into the adhA gene on pWUR104 by PCR amplification with primers BG417 (sense, 5′-CTTTAAGAAGGAGATATACCATG; creates a NcoI restriction site in PCR product) and BG1740 (antisense, 5′-GGAGCTCGAATTCGGATCCTCA; BamHI restriction site underlined). The error-prone PCR was conducted with REDTaq DNA polymerase under mutagenic reaction conditions (50 μl reaction mixture containing 10 ng template DNA, 200 ng primer BG417, 200 ng primer BG1740, 0.2 mM dATP and dGTP, 1 mM dCTP and dTTP, 0.5 mM MnCl2 and 5 units REDTaq DNA polymerase) and by using the Genemorph random mutagenesis kit with the Mutazyme DNA polymerase (reaction conditions according to the manufacturer; Stratagene).
The resulting PCR products were digested with NcoI-BamHI and subsequently cloned into expression vector pET24d. The resulting plasmids were used to transform E. coli XL1-Blue. Transformation mixtures were plated onto selective Luria-Bertani agar plates containing 50 μg/ml kanamycin and incubated overnight at 37°C. Next, the mutant library was harvested from each Luria-Bertani agar plate by adding 1.5 ml Luria-Bertani medium and by gently resuspending the colonies from the plate. The QIAprep spin miniprep kit (Qiagen) was used for the isolation of plasmid and subsequently plasmid of the mutant library was transformed to E. coli BL21(DE3) harboring the tRNA helper plasmid pSJS1244. Transformation mixtures were plated onto selective Luria-Bertani agar plates containing 50 μg/ml kanamycin and 50 μg/ml spectinomycin. Single colonies were transferred to microtiter plates containing 200 μl/well Luria-Bertani medium supplemented with 10% (w/v) glycerol, 50 μg/ml kanamycin and spectinomycin. In each microtiter plate three reference wells were present: (1) E. coli BL21(DE3) harboring pSJS1244 and pWUR104 (expression of wild-type AdhA), (2) E. coli BL21(DE3) harboring pSJS1244 and an empty pET24d vector, (3) blank (not inoculated).
Screening for increased activity on 2,5-hexanedione at 30°C
A replica of the random AdhA library was prepared in microtiter plates containing 200 μl/well Luria-Bertani medium supplemented with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), 50 μg/ml kanamycin and spectinomycin. After overnight growth at 37°C, the E. coli cells were harvested by centrifugation at 4,000 rpm for 10 min in a Hermle 383 Z centrifuge and subsequently lysed by addition of 40 μl Bacterial Protein Extraction Reagent (B-PER; Pierce, Rockford, IL, USA). No negative effect of B-PER on AdhA activity was detected in control experiments. The lysed cells were centrifuged (4,000 rpm, 15 min) and 40 μl of the resulting supernatant (cell free extract) was transferred to a second set of microtiter plates containing 160 μl/well 0.1 M sodium phosphate (pH 7.0), 125 mM 2,5-hexanedione and 0.35 mM NADPH. The cell-free extract and assay buffer were mixed, incubated at 30°C and the absorbance at 340 nm was measured in each well using an iEMS reader MF (Thermo Electron Corporation, Breda, The Netherlands). Potential high-performance mutants were selected by a significant faster decrease of absorbance at 340 nm (due to conversion of NADPH to NADP+) compared to the wild-type enzyme. The temperature-dependent spontaneous degradation of NADPH was corrected for. These mutants were regrown in 5 ml Luria-Bertani medium supplemented with 50 μg/ml kanamycin and spectinomycin until a cell density of OD600 nm = 0.6 was reached. The culture was then induced with 0.1 mM IPTG and incubation of the culture at 37°C was continued overnight. Cells were harvested, resuspended in 20 mM Tris/HCl buffer (pH 7.8) and disrupted by sonication. The crude cell extract was centrifuged for 10 min at 15,000×g. The resulting supernatant (cell free extract) was heated for 30 min at 80°C and subsequently centrifuged for 10 min at 15,000×g. The resulting supernatant (heat stable cell free extract) was used to rescreen the potential high-performance mutants.
Production and purification of ADH
Five ml Luria-Bertani medium with kanamycin and spectinomycin (both 50 μg ml−1) was inoculated with E. coli BL21(DE3) harboring pSJS1244 and expression plasmids containing either wild-type or mutant adhA genes and incubated overnight in a rotary shaker at 37°C. Next, 1 ml of the preculture was used to inoculate 1 l Luria-Bertani medium with kanamycin and spectinomycin (both 50 mg l−1) in a 2-l conical flask and incubated in a rotary shaker at 37°C until a cell density of OD600 nm = 0.6 was reached. The culture was then induced with 0.2 mM IPTG and incubation of the culture was continued at 37°C for 18 h. Cells were harvested, resuspended in 20 mM Tris/HCl buffer (pH 7.5) and passed twice through a French press at 110 MPa. The crude cell extract was centrifuged for 20 min at 10,000×g. The resulting supernatant (cell free extract) was heated for 30 min at 80°C and subsequently centrifuged for 20 min at 10,000×g. The supernatant (heat stable cell free extract) was filtered (0.45 μm) and applied to a Q-sepharose high-performance (GE Healthcare, Chalfont, St Giles, UK) column (1.6 × 10 cm) equilibrated in 20 mM Tris/HCl buffer (pH 7.8). Proteins were eluted with a linear 560 ml gradient from 0.0 to 1.0 M NaCl, in the same buffer. Wild-type and potential high-performance mutants AdhA were produced and purified for further characterization.
SDS-PAGE electrophoresis
Protein composition was analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE, 10%) (Sambrook et al. 1989), using a Mini-Protean 3 system (Biorad, Hercules, CA, USA). Protein samples for SDS-PAGE were prepared by heating for 5 min at 100°C in the presence of sample buffer (0.1 M sodium phosphate buffer, 4% SDS, 10% 2-mercaptoethanol, 20% glycerol, pH 6.8). A broad range protein marker (Biorad) was used to estimate the molecular mass of the proteins.
Characterization of high-performance mutants
Activity assays
Rates of alcohol oxidation and aldehyde reduction were determined at 30°C, unless stated otherwise, by following either the reduction of NADP+ or the oxidation of NADPH at 340 nm using a Hitachi U2010 spectrophotometer, with a temperature-controlled cuvette holder. Each oxidation reaction mixture contained 50 mM glycine (pH 10.5), 125 mM of alcohol and 0.35 mM NADP+. The reduction reaction mixture contained 0.1 M sodium phosphate buffer (pH 7.0), 125 mM aldehyde or ketone and 0.35 mM NADPH. In all assays, the reaction was initiated by addition of an appropriate amount of enzyme. One unit of ADH was defined as the oxidation or reduction of 1 μmol of NADPH or NADP+ per min, respectively. Protein concentration was determined using Bradford reagents (Bio-Rad) with bovine serum albumin as a standard (Bradford 1976). The temperature-dependent spontaneous degradation of NADPH was corrected for.
Optimum temperature and thermostability
The thermostability of AdhA mutants (enzyme concentration: 0.36 mg ml−1 in 20 mM Tris buffer pH 7.8) was determined by measuring the residual activity (2,5-hexanedione reduction according to the standard assay) after incubation of a time series at 100°C. The temperature optimum was determined in 0.1 M sodium phosphate buffer pH 7.0 by analysis of initial rates of 2,5-hexanedione reduction in the range of 30–100°C.
Kinetics
The kinetic parameters K m and V max were calculated from multiple measurements (at least eight measurements, which were done in duplicate) using the Michaelis–Menten equation and the program Tablecurve 2D (version 5.0). All the reactions followed Michaelis-Menten type kinetics. The turnover number (k cat, s−1) was calculated as: V max × subunit molecular mass (26 kDa)/60.
DNA sequencing and three-dimensional structure analysis
Plasmid DNA was isolated from 5-ml cultures by using the QIAprep spin miniprep kit (Qiagen) and the sequence of the different clones was determined by sequence analysis of both DNA strands (BaseClear). Amino acid substitutions were deduced from identified mutations in the DNA sequence of the adhA gene. A structural model of AdhA was constructed by homology modeling using the crystal structures of the NADP+-dependent clavulanic acid dehydrogenase from Streptomyces clavuligerus as a template (PDB-ID: 2JAP and 2JAH) (MacKenzie et al. 2007). The primary sequence of this enzyme aligns with the complete AdhA sequence while sharing 30% sequence identity. The aligned sequences were modeled using Swiss Model (Schwede et al. 2003), an automated comparative protein-modeling server. The validity of the model was checked by WhatCheck (Hooft et al. 1996). The model was compared to several homologous short-chain ADH structures, i.e., to the sepiapterin reductase of Chlorobium tepidum (26% identity; CT0609; PDB-ID 2BD0) (Supangat et al. 2006), the gluconate-5-dehydrogenase of Thermotoga maritima (22% identity; TM0441; PDB-ID 1VL8), and the human 17-β-hydroxysteroid dehydrogenase (23% identity, PDB-ID 1A27) (Breton et al. 1996; Mazza et al. 1998).
Results
Construction of a random mutant AdhA library and screening for increased activity on 2,5-hexanedione at 30°C
The hyperthermostable short-chain alcohol dehydrogenase AdhA from P. furiosus is optimally active at 90°C and its activity at 30°C does not exceed 5% of its optimal activity. AdhA has activity on the substrate 2,5-hexanedione and reduces it stereoselectively to (2S,5S)-hexanediol. However, the activity on 2,5-hexanedione at 30°C, the preferred temperature for production of (2S,5S)-hexanediol in an industrial setting, is very low. To study the possibility of increasing this low activity at 30°C, while retaining the stability of the enzyme, AdhA was subjected to random mutagenesis and low-temperature activity screening. Random mutations were introduced into the adhA gene by error-prone PCR (Leung et al. 1989; Cadwell and Joyce 1992). The mutated adhA genes were cloned in an E. coli expression host and a library of approximately 1,500 clones was constructed. Complete DNA sequence analysis of nine randomly selected mutants revealed an average mutation frequency of 2.75 bp/adhA gene, indicating that on average two or three amino acids will be changed in each AdhA enzyme (data not shown).
The mutant AdhA library was screened for increased activity on 2,5-hexanedione at 30°C. This resulted in the identification of 30 mutants with significantly higher activity than the wild-type controls. Heat-stable cell free extracts of the 30 mutants were further analyzed for increased low-temperature activity on 2,5-hexanedione. Eventually, the seven most active mutants (harboring plasmids pWUR314–pWUR320) were selected. Wild-type and high-performance mutants AdhA were produced and purified for further characterization.
Characterization of high-performance mutants
To gain more insight into the performance of the mutants at low and high temperatures their activity with different substrates was tested at 30 and 70°C. The AdhA mutant 13F2 performed the best at 30°C with all substrates (Table 1). The maximum specific activity of 13F2 with 2,5-hexanedione at 30°C was tenfold higher compared to the activity of the wild-type enzyme. At 70°C the wild-type enzyme and mutant 13F2 performed the best. At this temperature the other high-performance mutants showed a decreased activity with the tested substrates. All mutants, with the exception of 16H2, also had an increased activity with 2-propanol at 30°C, which is important for the cofactor regeneration system. Previous experiments have shown that wild-type AdhA can perform substrate-coupled NADPH regeneration making use of the oxidation of 2-propanol to acetone (Fig. 1). Further experiments with 2-propanol showed that all mutants had retained their activities in the presence of an excess of 2-propanol (data not shown).
The temperature optimum was determined for the mutants 13F2, 5E12 and 6G6, the three mutants with the highest activity at 30°C with 2,5-hexanedione, and the wild-type enzyme. The temperature optimum of the wild-type enzyme was determined to be between 90 and 100°C, which is comparable to the optimum of 90°C found previously (Van der Oost et al. 2001). The temperature optimum of the three mutants is not changed (90°C), but their activity at lower temperature (60°C and below) is clearly increased when compared to the wild-type enzyme. Figure 2 shows the specific activity at 30 and 70°C of the three mutants (13F2, 5E12 and 6G6) and the wild-type enzyme.
Next, the stability of the best performing mutant, 13F2, was compared with the wild-type enzyme. Therefore, the thermostability of both enzymes was tested at 100°C. The wild-type enzyme had an extremely high resistance to thermal inactivation, which was shown by a half-life value of 178 min. The stability of the wild-type enzyme is increased when compared to previously determined values (Van der Oost et al. 2001). This is most likely due to the use of a higher enzyme concentration. The mutant 13F2 with a half-life value of 44 min was clearly less stable than the wild-type enzyme, but it still exhibits a very high resistance to thermal inactivation.
The kinetic properties at 70°C of wild-type enzyme and mutant 13F2 were determined for the substrates 2,5-hexanedione, pyruvaldehyde, (2S.5S)-hexanediol and 2-pentanol, as well as for the cofactors used in these reactions (Table 2). The wild-type enzyme showed a higher affinity for all substrates and mutant 13F2 had merely a higher affinity for the cofactor NADPH, which is used in the reduction reaction.
Amino acid substitutions in high-performance mutants
As with all members of the short-chain dehydrogenase/reductase (SDR) family, AdhA from P. furiosus contains a well conserved NADP(H)-binding site at the N-terminus (Gly8-X-X-X-Gly12-X-Gly14), as well as several residues that are directly or indirectly involved in catalysis, including Asp59, Asn110, Ser137, Tyr150, Lys154, Thr183 (Van der Oost et al. 2001; Filling et al. 2002). A three-dimensional model of AdhA was constructed using the crystal structure of the NADP+-dependent clavulanic acid dehydrogenase from Streptomyces clavuligerus (MacKenzie et al. 2007).
The seven most active mutants were sequenced and amino acid substitutions were deduced from the changes in the DNA sequence (Table 3). This revealed that the high-performance mutants (13F2, 5E12 and 6G6) contained one or two amino acid substitutions. The mutant 13F2 has the amino acid substitution R11L, which is located in the NADP(H)-binding site (Fig. 3). Although the mutation is not at a conserved position of the NADP(H)-binding motif, the kinetic data support the hypothesis that the mutation enhances cofactor-binding affinity and turnover rate, as shown by a lowered K m and increased V max. Moreover, according to Wierenga et al. (1986) a hydrophobic residue such as leucine is preferred at this position in the NADP(H)-binding site. The second substitution of mutant 13F2, A180V, is located in the substrate-binding pocket near the conserved Thr183, which is involved in catalysis (Fig. 3). A valine at this position is also observed in several closely related AdhA homologs from mesophilic organisms. Mutation L176P is observed in two independent mutants (5E12 and 4A2) and the residue is located in the β-strand preceding an active site loop containing Thr183 (Fig. 3). Proline is most commonly found in turns, loops, the edge strands of β-sheets and the beginning or end of α-helices. Possibly this substitution is involved in a repositioning of the subsequent active site loop. While the introduction of proline residues at certain positions (in loop regions) has been associated with increased (thermo)stability and rigidification (Vieille and Zeikus 2001), in AdhA it appears to enhance low-temperature activity. The amino acid substitution in mutant 6G6, T153A, is located near the residues Tyr150 and Lys154, which are both members of the conserved catalytic tetrad in short-chain alcohol dehydrogenases (Fig. 3) (Filling et al. 2002). This mutation appears to have a conformational effect close to the active site, which is supported by the observation that this mutant can accept more bulky substrates than the wild-type enzyme (T. Dauβmann, personal communication). Mutation N86D that occurs in mutant 16H2 is also located in the cofactor-binding site, adding negative charge to the bottom of the pocket. Finally, the mutation R213I and K209T that occur in two different mutants (16H2 and 17A3) are spatially very close in the tertiary structure. This suggests that the enhanced activity of these mutants at low-temperature is due to a loss of positive charge at this site. Interestingly, close inspection of the model reveals two carboxyl residues (E23 and D24) in the C-terminal part of the neighboring α-helix within ion pair forming range of the R213 and K209. Most likely the increased activity at low-temperature is due to the loss of this ionic interaction, which possibly makes the enzyme more flexible at lower temperatures. This observation correlates well with the increased occurrence of ion pairs in proteins from hyperthermophiles compared to mesophiles (Karshikoff and Ladenstein 2001). The stabilizing feature is apparently rapidly lost when the selection pressure for extreme thermostability is relieved, and when there is a screening for low-temperature activity.
Ribbon representation of the AdhA structural model based on the clavulanic acid dehydrogenase (PDB-ID 2JAP and 2JAH) from Streptomyces clavuligerus (MacKenzie et al. 2007). Mutated amino acids and the NADP+-cofactor are indicated as sticks
Discussion
Our results show that the probed sequence space in the AdhA library was sufficient and the screening method suitable to detect mutants with an increase in low-temperature activity. The high-performance mutant 13F2 has a tenfold increase in low-temperature activity on 2,5-hexanedione. Its stability compared to the wild-type enzyme has been slightly compromised, but it still exhibits high resistance to thermal inactivation. The decrease in stability could be the result of increased flexibility, which might be the reason for enhanced low-temperature activity (Vieille and Zeikus 2001; Liang et al. 2004; Shiraki et al. 2001). It was also shown that the substrate coupled NADPH regeneration using 2-propanol could still be used.
The amino acid substitutions in the most enhanced mutants revealed that there are several adaptations in different regions of the enzyme, which can enhance the low-temperature activity. Regions of the enzyme where the mutations were found include the substrate-binding pocket, the cofactor-binding site and near the catalytic site. Lebbink et al. (2000) reported a similar pattern when they improved the low-temperature catalysis of the β-glucosidase (CelB) from P. furiosus. Characterization of the latter CelB mutants showed that the increase in low-temperature activity was achieved in different ways, including altered substrate specificity and increased flexibility.
In conclusion, the laboratory evolution approach has again been successful in engineering an improved enzyme. The best mutant (13F2) has an increased low-temperature activity, retained a high level of stability and can still use the efficient cofactor regeneration process, substrate coupled NADPH regeneration, at the lower temperature. This high-performance mutant can be used for the enzymatic conversion of 2,5-hexanedione to (2S,5S)-hexanediol or as a new starting point for further improvement.
References
Bastos FM, Dos Santos AG, Jones J, Oestreicher EG, Pinto GF, Paiva LMC (1999) Three different coupled enzymatic systems for in situ regeneration of NADPH. Biotechnol Tech 13:661–664
Bornscheuer UT, Pohl M (2001) Improved biocatalysts by directed evolution and rational protein design. Curr Opin Chem Biol 5:137–143
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Breton R, Housset D, Mazza C, Fontecilla-Camps JC (1996) The structure of a complex of human 17-β-hydroxysteroid dehydrogenase with estradiol and NADP+ identifies two principal targets for the design of inhibitors. Structure 4:905–915
Brunel JM, Faure B (2004) Enantioselective palladium catalyzed allylic substitution with a new phosphite ligand issued from (2S,5S)-hexanediol. J Mol Catal A Chem 212:61–64
Cadwell RC, Joyce GF (1992) Randomization of genes by PCR mutagenesis. PCR Methods Appl 2:28–33
Eckstein M, Dauβmann T, Kragl U (2004) Recent Developments in NAD(P)H regeneration for enzymatic reductions in one- and two-phase systems. Biocatal Biotransform 22:89–96
Fiala G, Stetter KO (1986) Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch Microbiol 145:56–61
Filling C, Berndt KD, Benach J, Knapp S, Prozorovski T, Nordling E, Ladenstein R, Jörnvall H, Oppermann U (2002) Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J Biol Chem 277:25677–25684
Haberland J, Hummel W, Daussmann T, Liese A (2002a) New continuous production for enantiopure (2R,5R)-hexanediol. Org Process Res Dev 6:458–462
Haberland J, Kriegesmann A, Wolfram E, Hummel W, Liese A (2002b) Diastereoselective synthesis of optically active (2R,5R)-hexanediol. Appl Microbiol Biotechnol 58:595–599
Hooft RW, Vriend G, Sander C, Abola EE (1996) Errors in protein structures. Nature 381(6580):272
Hummel W (1999) Large-scale applications of NAD(P)-dependent oxidoreductases: recent developments. Trends Biotechnol 17:487–492
Karshikoff A, Ladenstein R (2001) Ion pairs and the thermotolerance of proteins from hyperthermophiles: a “traffic rule” for hot roads. Trends Biochem Sci 26:550–556
Kim R, Sandler SJ, Goldman S, Yokota H, Clark AJ, Kim SH (1998) Overexpression of archaeal proteins in Escherichia coli. Biotechnol Lett 20:207–210
Lebbink JHG, Kaper T, Bron P, Van der Oost J, De Vos WM (2000) Improving low-temperature catalysis in the hyperthermostable Pyrococcus furiosus β-glucosidase CelB by directed evolution. Biochemistry 39:3656–3665
Leung DW, Chen E, Goeddel DV (1989) A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique 1:11–15
Liang ZX, Tsigos I, Bouriotis V, Klinman JP (2004) Impact of protein flexibility on hydride-transfer parameters in thermophilic and psychrophilic alcohol dehydrogenases. J Am Chem Soc 126:9500–9501
Lieser JK (1983) A simple synthesis of (S,S)-2,5-hexanediol. Synth Commun 13:765–767
MacKenzie AK, Kershaw NJ, Hernandez H, Robinson CV, Schofield CJ, Andersson I (2007) Clavulanic acid dehydrogenase: structural and biochemical analysis of the final step in the biosynthesis of the beta-lactamase inhibitor clavulanic acid. Biochemistry 46:1523–1533
Mazza C, Breton R, Housset D, Fontecilla-Camps JC (1998) Unusual charge stabilization of NADP+ in 17-β-hydroxysteroid dehydrogenase. J Biol Chem 273:8145–815
Radianingtyas H, Wright PC (2003) Alcohol dehydrogenases from thermophilic and hyperthermophilic archaea and bacteria. FEMS Microbiol Rev 27:593–616
Reid MF, Fewson CA (1994) Molecular characterization of microbial alcohol dehydrogenases. Crit Rev Microbiol 20:13–56
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Schmid A, Dordick JS, Hauer B, Kiener A, Wubbolts M, Witholt B (2001) Industrial biocatalysis today and tomorrow. Nature 409:258–268
Schoemaker HE, Mink D, Wubbolts MG (2003) Dispelling the myths - biocatalysis in industrial synthesis. Science 299:1694–1697
Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31:3381–3385
Shiraki K, Nishikori S, Fujiwara S, Hashimoto H, Kai Y, Takagi M, Imanaka T (2001) Comparative analyses of the conformational stability of a hyperthermophilic protein and its mesophilic counterpart. Eur J Biochem 268:4144–4150
Sorensen HP, Sperling-Petersen HU, Mortensen KK (2003) Production of recombinant thermostable proteins expressed in Escherichia coli: completion of protein synthesis is the bottleneck. J Chromatogr B Analyt Technol Biomed Life Sci 786:207–214
Supangat S, Seo KH, Choi YK, Park YS, Son D, Han CD, Lee KH (2006) Structure of Chlorobium tepidum sepiapterin reductase complex reveals the novel substrate binding mode for stereospecific production of l-threo-tetrahydrobiopterin. J Biol Chem 281:2249–2256
Van der Oost J, Voorhorst WG, Kengen SWM, Geerling ACM, Wittenhorst V, Gueguen Y, De Vos WM (2001) Genetic and biochemical characterization of a short-chain alcohol dehydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. Eur J Biochem 268:3062–3068
Vieille C, Zeikus GJ (2001) Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol Rev 65:1–43
Wierenga RK, Terpstra P, Hol WG (1986) Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol 187:101–107
Yuan L, Kurek I, English J, Keenan R (2005) Laboratory-directed protein evolution. Microbiol Mol Biol Rev 69:373–392
Zeikus JG, Vieille C, Savchenko A (1998) Thermozymes: biotechnology and structure-function relationships. Extremophiles 2:179–183
Acknowledgments
This work was supported by the EU 5th framework program PYRED (QLK3-CT-2001-01676).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by F. Robb.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Machielsen, R., Leferink, N.G.H., Hendriks, A. et al. Laboratory evolution of Pyrococcus furiosus alcohol dehydrogenase to improve the production of (2S,5S)-hexanediol at moderate temperatures. Extremophiles 12, 587–594 (2008). https://doi.org/10.1007/s00792-008-0164-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-008-0164-8