Abstract
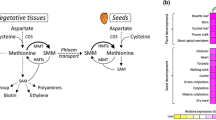
Methionine is a nutritionally essential sulfur-containing amino acid found at low levels in plant tissues. Yet, the factors that regulate its synthesis and accumulation in seeds are not fully known. Recent genetic studies demonstrate that Arabidopsis seeds are able to synthesize methionine de novo through the aspartate family pathway similarly to vegetative tissues; however, additional biochemical studies suggest that the S-methylmethionine (SMM) cycle also plays a major role in methionine synthesis in seeds. To better understand the contribution of these two pathways to methionine synthesis, we have sampled various vegetative and reproductive tissues during the Arabidopsis life cycle and determined the contents of soluble and protein-incorporated methionine, SMM, as well as the expression levels of the key genes involved in these two pathways. Our results strengthen the hypothesis that SMM that is produced in the rosette leaves from methionine contributes to methionine accumulation in seeds. However, the SMM cycle may have additional functions in plant tissues since its key genes were expressed in all of the examined tissues, although at different rates. The accumulation patterns of soluble and protein-incorporated methionine during the Arabidopsis life cycle were found to be similar to most of the other amino acids, especially to those belonging to the branched-chain and aromatic amino acids that are produced in chloroplasts together with methionine. This indicates that similar factors regulate the levels of amino acids during development.







Similar content being viewed by others
References
Amir R (2010) Current understanding of the factors regulating methionine content in vegetative tissues of higher plants. Amino Acids 39(4):917–931. doi:10.1007/s00726-010-0482-x
Amir R, Han T, Ma F (2012) Bioengineering approaches to improve the nutritional values of seeds by increasing their methionine content. Mol Breed 29:915–924
Bartlem D, Lambein I, Okamoto T, Itaya A, Uda Y, Kijima F, Tamaki Y, Nambara E, Naito S (2000) Mutation in the threonine synthase gene results in an over-accumulation of soluble methionine in Arabidopsis. Plant Physiol 123(1):101–110
Bourgis F, Roje S, Nuccio ML, Fisher DB, Tarczynski MC, Li C, Herschbach C, Rennenberg H, Pimenta MJ, Shen TL, Gage DA, Hanson AD (1999) S-methylmethionine plays a major role in phloem sulfur transport and is synthesized by a novel type of methyltransferase. Plant Cell 11(8):1485–1498
Bradbury LM, Ziemak MJ, Elbadawi-Sidhu M, Fiehn O, Hanson AD (2014) Plant-driven repurposing of the ancient S-adenosylmethionine repair enzyme homocysteine S-methyltransferase. Biochem J. doi:10.1042/BJ20140753
Cohen H, Israeli H, Matityahu I, Amir R (2014) Seed-specific expression of a feedback-insensitive form of cystathionine-gamma-synthase in arabidopsis stimulates metabolic and transcriptomic responses associated with desiccation stress. Plant Physiol 163(3):1575–1592. doi:10.1104/pp.114.246058
Curien G, Ravanel S, Dumas R (2003) A kinetic model of the branch-point between the methionine and threonine biosynthesis pathways in Arabidopsis thaliana. Eur J Biochem 270(23):4615–4627
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139(1):5–17. doi:10.1104/pp.105.063743
Galili G, Amir R (2013) Fortifying plants with the essential amino acids lysine and methionine to improve nutritional quality. Plant Biotechnol J 11(2):211–222. doi:10.1111/pbi.12025
Gallardo K, Firnhaber C, Zuber H, Hericher D, Belghazi M, Henry C, Kuster H, Thompson R (2007) A combined proteome and transcriptome analysis of developing Medicago truncatula seeds: evidence for metabolic specialization of maternal and filial tissues. Mol Cell Proteomics 6(12):2165–2179. doi:10.1074/mcp.M700171-MCP200
Gepstein S, Sabehi G, Carp MJ, Hajouj T, Nesher MF, Yariv I, Dor C, Bassani M (2003) Large-scale identification of leaf senescence-associated genes. Plant J 36(5):629–642
Giovanelli JG (1987) Sulfur amino acids in plants: an overview. Methods Enzymol 143:419–428
Guyer D, Patton D, Ward E (1995) Evidence for cross-pathway regulation of metabolic gene expression in plants. Proc Natl Acad Sci USA 92(11):4997–5000
Hacham Y, Avraham T, Amir R (2002) The N-terminal region of Arabidopsis cystathionine gamma-synthase plays an important regulatory role in methionine metabolism. Plant Physiol 128(2):454–462. doi:10.1104/pp.010819
Hacham Y, Matityahu I, Schuster G, Amir R (2008) Overexpression of mutated forms of aspartate kinase and cystathionine gamma-synthase in tobacco leaves resulted in the high accumulation of methionine and threonine. Plant J 54(2):260–271. doi:10.1111/j.1365-313X.2008.03415.x
Hacham Y, Matityahu I, Amir R (2013) Light and sucrose up-regulate the expression level of Arabidopsis cystathionine gamma-synthase, the key enzyme of methionine biosynthesis pathway. Amino Acids 45(5):1179–1190. doi:10.1007/s00726-013-1576-z
Hanafy MS, Rahman SM, Nakamoto Y, Fujiwara T, Naito S, Wakasa K, Ishimoto M (2013) Differential response of methionine metabolism in two grain legumes, soybean and azuki bean, expressing a mutated form of Arabidopsis cystathionine gamma-synthase. J Plant Physiol 170(3):338–345. doi:10.1016/j.jplph.2012.10.018
Hesse H, Hoefgen R (2003) Molecular aspects of methionine biosynthesis. Trends Plant Sci 8(6):259–262 S1360138503001079
Inaba K, Fujiwara T, Hayashi H, Chino M, Komeda Y, Naito S (1994) Isolation of an Arabidopsis thaliana mutant, mto1, that overaccumulates soluble methionine. Plant Physiol 104(3):881–887 104/3/881 [pii]
Jander G, Joshi V (2010) Recent progress in deciphering the biosynthesis of aspartate-derived amino acids in plants. Mol Plant 3(1):54–65. doi:10.1093/mp/ssp104
Joshi V, Jander G (2009) Arabidopsis methionine gamma-lyase is regulated according to isoleucine biosynthesis needs but plays a subordinate role to threonine deaminase. Plant Physiol 151(1):367–378. doi:10.1104/pp.109.138651
Kim J, Leustek T (2000) Repression of cystathionine γ-synthase in Arabidopsis thaliana produces partial methionine auxotrophy and developmental abnormalities. Plant Sci 151:9–18. doi:10.1016/S0168-9452(99)00188-0
Kim J, Lee M, Chalam R, Martin MN, Leustek T, Boerjan W (2002) Constitutive overexpression of cystathionine gamma-synthase in Arabidopsis leads to accumulation of soluble methionine and S-methylmethionine. Plant Physiol 128(1):95–107
Kocsis MG, Ranocha P, Gage DA, Simon ES, Rhodes D, Peel GJ, Mellema S, Saito K, Awazuhara M, Li C, Meeley RB, Tarczynski MC, Wagner C, Hanson AD (2003) Insertional inactivation of the methionine s-methyltransferase gene eliminates the s-methylmethionine cycle and increases the methylation ratio. Plant Physiol 131(4):1808–1815. doi:10.1104/pp.102.018846
Lam HM, Coschigano KT, Oliveira IC, Melo-Oliveira R, Coruzzi GM (1996) The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Ann Rev Plant Physiol Plant Mol Biol 47:569–593. doi:10.1146/annurev.arplant.47.1.569
Lee M, Huang T, Toro-Ramos T, Fraga M, Last RL, Jander G (2008) Reduced activity of Arabidopsis thaliana HMT2, a methionine biosynthetic enzyme, increases seed methionine content. Plant J 54(2):310–320. doi:10.1111/j.1365-313X.2008.03419.x
Maeda H, Dudareva N (2012) The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu Rev Plant Biol 63:73–105. doi:10.1146/annurev-arplant-042811-105439
Matityahu I, Godo I, Hacham Y, Amir R (2013) Tobacco seeds expressing feedback-insensitive cystathionine gamma-synthase exhibit elevated content of methionine and altered primary metabolic profile. BMC Plant Biol 13:206. doi:10.1186/1471-2229-13-206
Mudd H, Datko H (1990) The S-methylmethionine cycle in Lemna paucicostata. Plant Physiol 93:623–630
Nikiforova VJ, Kopka J, Tolstikov V, Fiehn O, Hopkins L, Hawkesford MJ, Hesse H, Hoefgen R (2005) Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol 138(1):304–318
Ranocha P, McNeil SD, Ziemak MJ, Li C, Tarczynski MC, Hanson AD (2001) The S-methylmethionine cycle in angiosperms: ubiquity, antiquity and activity. Plant J 25(5):575–584 tpj988 [pii]
Ravanel S, Gakiere B, Job D, Douce R (1998) The specific features of methionine biosynthesis and metabolism in plants. Proc Natl Acad Sci USA 95(13):7805–7812
Roje S (2006) S-Adenosyl-l-methionine: beyond the universal methyl group donor. Phytochemistry 67(15):1686–1698. doi:10.1016/j.phytochem.2006.04.019
Song S, Hou W, Godo I, Wu C, Yu Y, Matityahu I, Hacham Y, Sun S, Han T, Amir R (2013) Soybean seeds expressing feedback-insensitive cystathionine γ-synthase exhibit a higher content of methionine. J Exp Bot 64(7):1917–1926. doi:10.1093/jxb/ert053
Soudry E, Ulitzur S, Gepstein S (2005) Accumulation and remobilization of amino acids during senescence of detached and attached leaves: in planta analysis of tryptophan levels by recombinant luminescent bacteria. J Exp Bot 56(412):695–702. doi:10.1093/jxb/eri054
Tan Q, Zhang L, Grant J, Cooper P, Tegeder M (2010) Increased phloem transport of S-methylmethionine positively affects sulfur and nitrogen metabolism and seed development in pea plants. Plant Physiol 154(4):1886–1896. doi:10.1104/pp.110.166389
Tegeder M, Rentsch D (2010) Uptake and partitioning of amino acids and peptides. Mol Plant 3(6):997–1011. doi:10.1093/mp/ssq047
Tzin V, Galili G (2010) New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol Plant 3(6):956–972. doi:10.1093/mp/ssq048
Xia J, Psychogios N, Young N, Wishart DS (2009) MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res 37(Web Server issue):W652–W660. doi:10.1093/nar/gkp356
Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS (2012) MetaboAnalyst 2.0–a comprehensive server for metabolomic data analysis. Nucleic Acids Res 40(Web Server issue):W127–W133. doi:10.1093/nar/gks374
Acknowledgments
We would like to thank Dr. Gidi Baum for his critical reading and valuable comments. This work was supported by the Israel Science Foundation (Grant 231-09).
Conflict of interest
The authors declare that they have no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
A. Frank and H. Cohen contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Frank, A., Cohen, H., Hoffman, D. et al. Methionine and S-methylmethionine exhibit temporal and spatial accumulation patterns during the Arabidopsis life cycle. Amino Acids 47, 497–510 (2015). https://doi.org/10.1007/s00726-014-1881-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-014-1881-1