Abstract
The leafhopper Macrosteles laevis, like other plant sap-feeding hemipterans, lives in obligate symbiotic association with microorganisms. The symbionts are harbored in the cytoplasm of large cells termed bacteriocytes, which are integrated into huge organs termed bacteriomes. Morphological and molecular investigations have revealed that in the bacteriomes of M. laevis, two types of bacteriocytes are present which are as follows: bacteriocytes with bacterium Sulcia and bacteriocytes with Nasuia symbiont. We observed that in bacteriocytes with Sulcia, some cells of this bacterium contain numerous cells of the bacterium Arsenophonus. All types of symbionts are transmitted transovarially between generations. In the mature female, the bacteria Nasuia, bacteria Sulcia, and Sulcia with Arsenophonus inside are released from the bacteriocytes and start to assemble around the terminal oocytes. Next, the bacteria enter the cytoplasm of follicular cells surrounding the posterior pole of the oocyte. After passing through the follicular cells, the symbionts enter the space between the oocyte and follicular epithelium, forming a characteristic “symbiont ball.”
Similar content being viewed by others
Introduction
Macrosteles laevis is a representative of the subfamily Deltocephalinae, the largest group within the family Cicadellidae (Zahniser and Dietrich 2010, 2013). The Deltocephalinae leafhoppers are distributed worldwide. Most of them feed on xylem sap, using a needle-like mouthpart. Due to their feeding habitat, many of these insects are vectors of economically important plant diseases (caused by phytopathogenic viruses, bacteria, or fungi) (Brčak 1979).
The Deltocephalinae leafhoppers, like other plant sap-feeding hemipterans, harbor obligate symbiotic microorganisms, which synthesize and provide them with the amino acids missing in their diet (Douglas 1998, 2003, 2006; Baumann 2005).
Buchner (1965), basing his work on histological studies, distinguished two groups of symbionts: primary symbionts (also termed P-symbionts) and accessory symbionts (also termed secondary or facultative or S-symbionts). The primary symbionts provide their host insect with essential nutrients; therefore, their presence in the host body is necessary for its survival (i.e., proper functioning and reproduction) (Douglas 1998, 2006; Baumann et al. 2006; Moran and Dale 2006). The primary symbiosis is a result of a single long ago infection of an ancestor of the insect group by a free-living microorganism (review: Baumann 2005, 2006; Baumann et al. 2006). In consequence, such microorganisms are present in all representatives of this insect group. The obligate symbionts may be bacteria or yeast. The primary symbionts of Hemiptera: Sternorrhyncha (aphids, scale insects, whiteflies, and psyllids), Hemiptera: Cicadomorpha, Hemiptera: Fulgoromorpha, Hemiptera: Coleorrhyncha, and in some heteropterans are harbored in giant cells of mesodermal origin termed bacteriocytes (if bacteria) or mycetocytes (if yeast) (Buchner 1965; Baumann 2005, 2006; Baumann et al. 2006; Kuechler et al. 2013). These microorganisms are transovarially (vertically) transmitted between generations (see Buchner 1965 for further details).
Secondary symbionts may be present only in some populations of the species (review: Baumann 2005). Their presence in the insect body is the result of a more recent infection of an ancestor. The secondary symbionts may occur in bacteriocytes or in other types of insect cells (e.g., fat body cells), or free in hemolymph. These symbionts are not only transmitted transovarially but also between specimens of the same population (horizontally). The function of the S-symbionts was the object of many recent studies (e.g., Montlor et al. 2002; Oliver et al. 2003; Scarborough et al. 2005) which have shown that they impart specific properties to insects, such as the ability to survive heat stress or attack of parasitic hymenopterans or pathogenic fungi. In comparison to other hemipterans, there is still little data on the symbiotic systems (i.e., types of symbionts, their ultrastructure, distribution in the body cavity of insects, and mode of transovarial transmission) of Cicadomorpha. Earlier histological observations (Buchner 1965; Müller 1962) and more recent ultrastructural and molecular analyses (Moran et al. 2003; Takiya et al. 2006; Bressan et al. 2009; Noda et al. 2012; Bennett and Moran 2013; Ishii et al. 2013; Koga et al. 2013; Michalik et al. 2014; Szklarzewicz et al. 2015) have shown that these hemipterans may be hosts to several types of obligate symbionts. Recent molecular analyses (Takiya et al. 2006; Wu et al. 2006; McCutcheon et al. 2009; McCutcheon and Moran 2010) have revealed that in contrast to Sternorrhyncha, in which only primary symbionts are engaged in the synthesis of amino acids, in Cicadomorpha, all symbionts (termed by Takiya et al. 2006 as “coprimary symbionts”) synthesize these substances. Molecular analyses have revealed that members of Cicadomorpha and Fulgoromorpha (both formerly treated as Auchenorrhyncha) usually harbor the obligate Bacteroidetes bacterium “Candidatus Sulcia muelleri” (hereafter Sulcia) and one other kind of the coprimary symbionts belonging to the phylum Proteobacteria, e.g., “Candidatus Baumannia cicadellinicola” (hereafter Baumannia), “Candidatus Zinderia insecticola” (hereafter Zinderia), “Candidatus Nasuia deltocephalinicola” (hereafter Nasuia), or “Candidatus Hodgkinia cicadicola” (hereafter Hodgkinia) (Takiya et al. 2006; Urban and Cryan 2012; Bennett and Moran, 2013; Ishii et al. 2013; Koga et al. 2013). According to Moran et al. (2005) and Koga et al. (2013), the common ancestor of the Cicadomorpha and Fulgoromorpha was infected by the bacterium Sulcia and betaproteobacterial symbiont over 260 Ma ago. During the further evolution of some lineages of these hemipterans, the betaproteobacterium has been replaced by other bacteria, e.g., gammaproteobacterium Baumannia in some Cicadellinae and alphaproteobacterium Hodgkinia in cicadas. In some planthopper families and Deltocephalinae leafhoppers, the bacterial symbionts have been replaced by yeast symbionts (Noda 1977; Marzorati et al. 2006; Sacchi et al. 2008; Michalik et al. 2009).
To date, little is known about the symbiotic systems of the Deltocephalinae leafhoppers; however, the results of a few previous studies (Buchner 1925, 1965; Müller 1962; Marzorati et al. 2006; Sacchi et al. 2008; Wangkeeree et al. 2011; Noda et al. 2012; Bennett and Moran 2013; Ishii et al. 2013) suggest that these hemipterans are characterized by a large diversity of their symbionts. The aim of this study was to characterize the symbiotic system of the widely distributed species, Macrosteles laevis, in the Holarctic region.
Material and methods
Insects
Adult males and females of M. laevis (Ribaut, 1927) (Fig. 1a) destined for histological and ultrastructural studies were collected from herbaceous plants in Kraków, Gliwice, Częstochowa, and Bielsko-Biała (in the south of Poland) from June to September during the years 2012–2014. Individuals destined for molecular analyses were collected in Częstochowa.
a Adult female of Macrosteles laevis. b Cross section through the abdomen of the adult female. Bacteriomes (encircled with black dotted line) are composed of two distinct types of bacteriocytes: peripherally located bacteriocytes with bacterium Sulcia (white asterisk) and centrally located bacteriocytes with bacterium Nasuia (black asterisk). Oocyte (oc). c Dissected bacteriomes. Stereomicroscope, scale bar = 0.5 mm
DNA isolation
The bacteriomes were dissected from about 80 adult females of M. laevis and fixed in 100 % ethanol. DNA was extracted using Sherlock AX extraction kit (A&A Biotechnology) according to the manufacturer’s protocol with the following modification: before DNA extraction, bacteriomes were shaken for 60 s using a Beadbeater device with the addition of 0.3 ml distillated water, 0.3 ml lysis buffer L1.4, 20 μl proteinase K, and 0.5 g zirconia beads. DNA isolated from bacteriocytes was stored at −20 °C for further analyses.
PCR, cloning, and sequencing
Bacterial 16S ribosomal DNA (rDNA) was amplified using the following primers: Late11B-all For: AGAAGGAGATATAACTAT GAG TTT GAT CCT GGC TCA G and Late11B-all Rev: GGAGATGGGAAGTCATTACG GCT ACC TTA CGA CTT. Next, PCR products were cloned into pLATE 11 plasmid vector using Thermo Scientific aLICator Ligation Independent Cloning and Expression System kit. The ligated mixture was transformed into competent cells Escherichia coli TOP10F which were prepared using E. coli Transformer kit (A&A Biotechnology). After 16 h, the occurrence of bacterial 16S rDNA was confirmed by a diagnostic PCR reaction with the following primers: pJET For: GCCTGAACACCATATCCATCC and pJET Rev: GCAGCTGAGAATATTGTAGGAGAT.
After restrictive analysis, the plasmids were isolated using Plasmid Mini Ax kit (A&A Biotechnology) and then six clones were sequenced. The determined nucleotide sequences were deposited in the GenBank database under the accession numbers KR337979–KR337981.
Phylogenetic analysis
The phylogenetic analyses of symbiotic microorganisms were performed using the obtained sequences of 16S rDNA and homologous sequences downloaded from the GenBank database. The sequences were edited using BioEdit Sequence Alignment Editor 5.0.9 (Hall 1999); next, the sequence alignments were generated using ClustalX 1.8 (Thompson et al. 1997). All of the alignments were verified and then visually corrected. For the analyses, the appropriate nucleotide substitution model was first determined using the Modeltest 3.06 (Posada and Crandall 1998). The variable positions of the obtained 16S rDNA sequences were calculated using DnaSP software, whereas the base compositions of all analyzed genes were estimated using MEGA 6.0. software (Tamura et al. 2013). Phylogenetic analyses were conducted using Beast v 1.6.1 (Drummond and Rambaut 2007). The time of lineage divergence was estimated using the uncorrelated lognormal relaxed clock model. For each matrix, Beast was run using a Yule speciation process and two independent MCMC chains for 10,000,000 generations, sampling every 100th generation. The convergence to stationary and the effective sample size of the model parameters were checked using Tracer. The maximum clade credibility trees were built with TreeAnnotator (Drummond and Rambaut 2007). FigTree 1.4.0 software (Rambaut 2008) was used to visualize the results of phylogenetic analyses.
Stereomicroscope observations
The specimens were fixed in 70 % ethanol. The adult specimens, sectioned abdomens, and dissected bacteriomes were photographed using a Nikon SMZ 1500 stereomicroscope.
Light and electron microscopy analyses
For histological and ultrastructural studies, entire abdomens and dissected ovaries from about 80 individuals were examined. The specimens were fixed in 2.5 % glutaraldehyde in 0.1 M phosphate buffer (pH 7.4), rinsed in the same buffer with the addition of sucrose (5.8 g/100 ml), and postfixed in 1 % osmium tetroxide in the same buffer. Next, the material was dehydrated in a graded series of alcohols and acetone and embedded in epoxy resin Epon 812 (Serva, Heidelberg, Germany). Semithin sections (1 μm thick) were stained with 1 % methylene blue in 1 % borax and examined under light microscopes Leica DMR and Nikon Eclipse 80i. Ultrathin sections (90 nm thick) were contrasted with uranyl acetate and lead citrate and examined in Jeol JEM 2100 (Jeol, Japan) electron transmission microscope (TEM) at 80 kV.
Results
Molecular identification of symbiotic microorganisms of M. laevis and their phylogenetic analysis
Restrictive analysis and sequencing of the bacterial 16S rDNA clearly indicated the occurrence of three types of clones in the bacteriomes of M. laevis. One of them (signed as type 1) was dominant, whereas the other two types (signed as types 2 and 3) were less numerous. The nucleotide sequences obtained were analyzed and compared with the GenBank data using Blast. The Blast searches clearly demonstrated the affiliation of clones of type 1 to “Candidatus Sulcia muellerii” (Bacteroidetes) (99 % identity with 16S rDNA sequence of Sulcia symbiont of Macrosteles quadrilineatus [CP006060]. Type 2 clones were identified as a “Candidatus Nasuia deltocephalinicola” (Betaproteobacteria), with a top Blast hit to 16S rDNA sequence of Nasuia symbiont isolated from Macrosteles sexnotatus [AB795339]. Type 3 clones displayed the highest similarity to Arsenophonus symbiont of an aphid Aphis melosae [KF824523] and a scale insect Ericerus pela [JN990929].
The variation within the 16S rDNA sequences used in phylogenetic analyses was about 12 % variable sites and 8.5 % parsimony informative sites for Sulcia symbionts and about 47.5 % variable sites and 39.5 % parsimony informative sites for the Nasuia ones. The mean base compositions of the Sulcia symbionts were as follows: 30.9 % A, 25.1 % T, 17.3 % C, and 26.6 % G, while in the Nasuia symbionts, there were 33.9 % A, 26.8 % T, 14.9 % C, and 24.3 % G.
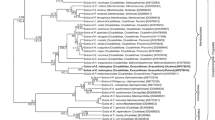
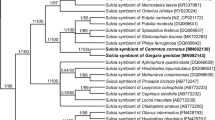
The phylogenetic analysis of 16S rDNA sequences of Sulcia symbionts of various representatives of Cicadomorpha confirmed the affiliation of type 1 clones to Bacteroidetes symbionts. The phylogenetic tree (Fig. 2) indicates that the Sulcia symbionts of members of the Deltocephalinae subfamily create a monophyletic group within the symbionts of Membracoidea with 1.00 posterior probability values. The result of the phylogenetic analysis of 16S rDNA sequences from proteobacterial symbionts of the members of Cicadomorpha (Fig. 3) generally matches up with Blast search results. It was indicated that among proteobacterial symbionts of Cicadomorpha, Nasuia symbionts of representatives of Deltocephalinae subfamily are closely related and form a monophyletic group.
Ultrastructure and distribution of symbiotic microorganisms
Ultrastructural studies on symbionts of M. laevis revealed that within the body of each individual, three types of intracellular symbiotic bacteria occur. All the symbionts are harbored in the cytoplasm of bacteriocytes (Fig. 4a, b). Bacteriocytes form two large, yellow-colored structures termed bacteriomes that occur between the body wall and the gonads (Fig. 1b, c). The bacteriomes are surrounded by a thin sheath composed of a monolayered epithelium. These organs are present in both males and females; however, in females, they are much larger. Two types of bacteriocytes may be distinguished within each bacteriome: (1) the peripherally located bacteriocytes (Fig. 1b) filled with large, pleomorphic bacteria which stain intensely with methylene blue (Fig. 4a, b) and are electron-dense under an electron transmission microscope (Fig. 4c, d) and (2) the centrally located bacteriocytes (Fig. 1b) containing pleomorphic bacteria that stain with methylene blue less intensely (Fig. 4b) and are electron-translucent under an electron transmission microscope (Fig. 4e). Within the outer part of the bacteriome, in the cytoplasm of some large, pleomorphic bacteria, numerous long, rod-shaped bacteria occur (Fig. 4a, b, d).
Organization of the bacteriome of Macrosteles laevis. a Fragment of the bacteriome comprising the bacteriocytes with bacterium Sulcia (white arrows) and bacterium Sulcia containing bacterium Arsenophonus (black arrows). Bacteriome sheath (bs). Methylene blue, scale bar = 20 μm. b Fragment of the bacteriome with bacteriocytes containing the bacterium Sulcia (white arrows), bacterium Sulcia with bacterium Arsenophonus inside (black arrows), and bacteriocytes with betaproteobacterial symbiont Nasuia (black arrowheads). Bacteriocyte nucleus (bn). Methylene blue, scale bar = 20 μm. c Fragment of the bacteriocyte filled with bacterium Sulcia (S). TEM, scale bar = 2 μm. d Fragment of the bacteriocyte filled with bacterium Sulcia (S) containing the bacterium Arsenophonus (white arrowheads). TEM, scale bar = 2 μm. e Fragment of the bacteriocyte filled with Nasuia (N). Bacteriocyte nucleus (bn). TEM, scale bar = 2 μm
The symbiotic bacteria present in the bacteriomes of M. laevis were identified based on the comparison of histological, ultrastructural, and molecular results. Based on the ultrastructural features of bacteria Sulcia and Nasuia (Bressan et al. 2009; Noda et al. 2012; Michalik et al. 2014; Szklarzewicz et al. 2015), the electron-dense pleomorphic bacteria localized in the peripheral part of the bacteriome have been identified as the Bacteroidetes species Sulcia, whereas electron-translucent pleomorphic bacteria occupying the inner part of the bacteriome as the betaproteobacterial symbiont Nasuia. The third type of symbionts, i.e., long, rod-shaped bacterium occurring inside the cells of Sulcia, has been identified as the gammaproteobacterial species Arsenophonus.
Transovarial transmission of symbiotic microorganisms
All types of symbionts in the species studied are transovarially transmitted from mother to the offspring. The description of the consecutive stages of transovarial transmission of symbiotic bacteria was possible because the specimens of M. laevis were collected and fixed from June to September.
The ovaries of M. laevis are composed of seven ovarioles of telotrophic type (for further details concerning the classification and organization of insect ovarioles, see Büning 1994; Biliński 1998) that via ovariolar stalks (pedicels) join with lateral oviduct. Each ovariole contains several linearly arranged oocytes that are surrounded by a single layer of follicular cells (Figs. 5a and 6a). In the females containing ovaries with terminal oocytes in the stage of early vitellogenesis, bacteria Nasuia, bacteria Sulcia, and bacteria Sulcia filled with bacteria Arsenophonus leave the cytoplasm of bacteriocytes and begin to invade ovarioles. Before migrating from the bacteriocytes into the gonads, the bacteria change their shape from irregular into roughly spherical (Fig. 5a, e). The symbiotic microorganisms enter the follicular cells in the region of the posterior pole of terminal oocytes (Figs. 5a and 6b) and temporarily accumulate in their cytoplasm (Figs. 5b–e and 6b). During this invasion of symbionts, the volume of infected follicular cells significantly increases (Figs. 5b, d and 6b). Next, the bacteria leave the follicular cells and enter the perivitelline space (i.e., space between the oocyte and follicular epithelium) (Figs. 5d and 6b). The symbionts assemble in the invagination of the oolemma of the posterior pole of the oocyte initially in the lens-like form (Figs. 5d and 6b) and finally in the spherical form termed “symbiont ball” (Figs. 5f and 6c). The bacteria residing in the “symbiont ball” change their shape from spherical to polygonal and closely adhere to one another (Figs. 5d–f and 6c). When the infection of the ovariole is near termination, the number of organelles in the cytoplasm of follicular cells of the posterior pole of the oocyte, especially the cisternae of rough endoplasmic reticulum and Golgi complexes, increases greatly (Fig. 5e). These changes in the ultrastructure of follicular cells are associated with the production of egg envelopes.
Consecutive stages of the infectioning of the terminal oocyte of the ovariole by symbiotic bacteria. a, b Posterior pole of the terminal oocyte (oc) (a early vitellogenic oocyte; b late vitellogenic oocyte) surrounded by a single layer of follicular cells (fc) (longitudinal section). Symbiotic bacteria enter the cytoplasm of follicular cells. Sulcia (white arrows), Sulcia with bacterium Arsenophonus inside (black arrows), Nasuia (black arrowheads), nucleus of follicular cell (fn), ovariolar stalk (pedicel) connecting the ovariole to the lateral oviduct (p). Methylene blue, scale bar = 20 μm. c High-magnification view of the follicular cell during the transmission of symbiotic bacteria. Sulcia (S), Arsenophonus (white arrowheads), Nasuia (N). TEM, scale bar = 2 μm. d Fragment of the terminal oocyte (oc) and enlarged follicular cells (encircled with black dotted line) filled with symbiotic bacteria. Note the lens-shaped accumulation of the bacteria in the perivitelline space (encircled with white dotted line) (longitudinal section). Follicular cells (fc), Sulcia (white arrows), Sulcia with bacterium Arsenophonus inside (black arrows), Nasuia (black arrowheads). Methylene blue, scale bar = 20 μm. e High-magnification view of follicular cell (fc) filled with symbiotic bacteria and perivitelline space (black double arrows) with the accumulation of symbionts. Sulcia (S), Arsenophonus (white arrowheads), Nasuia (N), nucleus of follicular cell (fn), cisternae of rough endoplasmic reticulum (rer). TEM, scale bar = 2 μm. f The accumulation of symbionts (“symbiont ball”) in the perivitelline space in the deep invagination of oolemma. Oocyte (oc), Sulcia (white arrows), Sulcia with bacterium Arsenophonus inside (black arrows), Nasuia (black arrowheads). Methylene blue, scale bar = 20 μm
Schematic representation of the transovarial transmission of symbionts in Macrosteles laevis. a The terminal oocytes are accompanied by bacteriocytes. b Symbiotic bacteria leave the bacteriocytes, invade follicular cells surrounding posterior pole of the terminal oocyte, and enter the space between the oocyte and follicular epithelium. c Symbiotic bacteria accumulate in the deep invagination of the oolemma forming a characteristic “symbiont ball.” Symbiotic bacteria (s), bacteriocyte (bc), follicular cells (fc), oocyte (oc), oocyte nucleus (n), “symbiont ball” (sb)
Discussion
Müller (1962) and Buchner (1965), on the basis of histological observations of several species of the Deltocephalinae, distinguished two types of their symbionts termed a-symbionts and t-symbionts. Results of recent molecular studies on symbionts living in several species belonging to three genera, Macrosteles, Nephotettix, and Matsumuratettix, have shown that they harbor the bacterium Sulcia and the betaproteobacterial symbiont Nasuia which correspond to a-symbionts and t-symbionts (respectively) (Wangkeeree et al. 2011; Noda et al. 2012; Bennett and Moran 2013; Ishii et al. 2013; this study). Thus, the above facts indicate that Macrosteles, Nephotettix, and Matsumuratettix retained the ancestral dual symbiotic system (see “Introduction”). Molecular phylogenetic analyses (see Figs. 2 and 3) have revealed that both Sulcia and betaproteobacterial symbiont Nasuia living in M. laevis form well-defined clades with Sulcia and betaproteobacterial symbionts of other Deltocephalinae leafhoppers.
In contrast to the dual “Sulcia-betaproteobacteria” system, the bacteriocyte-associated bacterium Cardinium (Bacteroidetes) and mycetocyte-associated yeast-like symbionts are present in another member of the Deltocephalinae, Scaphoideus titanus (Marzorati et al. 2006; Sacchi et al. 2008). Furthermore, in the body of some Deltocephalinae leafhoppers, additional bacteria, e.g., Wolbachia, Rickettsia, Burkholderia, and Diplorickettsia, have been identified (Noda et al. 2012; Ishii et al. 2013). Our ultrastructural and molecular analyses have indicated that in M. laevis besides Sulcia and Nasuia, inside Sulcia cells, numerous gammaproteobacteria Arsenophonus are present. Thus, the state of symbiosis in the representatives of the Deltocephalinae appeared even more complex.
The presence of Arsenophonus inside the cells of the bacterium Sulcia is of special interest because such “an internal symbiosis” is very rare phenomenon within insects. To date, “symbionts within symbionts” have been reported only for some members of the scale insect family Pseudococcidae (mealybugs) (von Dohlen et al. 2001; Thao et al. 2002; Kono et al. 2008; Gatehouse et al. 2011; McCutcheon and von Dohlen 2011) and for the green leafhopper, Cicadella viridis (Cicadomoropha, Cicadellidae: Cicadellinae) (Michalik et al. 2014). Molecular analyses have revealed that in both mealybugs and in C. viridis, the internalized bacteria are closely related to the gammaproteobacterium Sodalis glossinidius that is the secondary symbiont of tsetse fly (Gatehouse et al. 2011; Michalik et al. 2014). In contrast to the closely related internal symbionts of mealybugs and C. viridis, their host bacteria are characterized by a different systematic affinity: in mealybugs, they belong to the class Betaproteobacteria of the phylum Proteobacteria (von Dohlen et al. 2001; Baumann et al. 2002; Gatehouse et al. 2011), and in C. viridis, to the phylum Bacteroidetes (Michalik et al. 2014). Additionally, in mealybugs, Sodalis-like bacteria are always harbored in the cytoplasm of betaproteobacterial symbionts, whereas in C. viridis, they may occur individually in the cytoplasm of their own bacteriocytes or may coreside in bacteriocytes with the bacterium Sulcia. It has been observed that during the coexistence of both bacteria in the cytoplasm of the same bacteriocyte, Sodalis-like bacteria enter the cytoplasm of Sulcia. In consequence, bacteria Sulcia containing internalized Sodalis-like bacteria are transmitted to the next generation. On the basis of the above observations, Michalik et al. (2014) suggested that the presence of Sodalis-like bacteria represents the youngest stage of association (in statu nascendi). Our ultrastructural observations have revealed that in M. laevis, as in mealybugs, the internalized bacteria never occur individually, but always (i.e., in bacteriocytes, during the infectioning of ovaries, and in the ovarioles) live inside the cells of the bacterium Sulcia. This fact suggests that just as in the case of mealybugs (Thao et al. 2002), the association of both symbionts and M. laevis is of more ancient origin than in C. viridis.
The gammaproteobacterium Arsenophonus is widespread within arthropods (Nováková et al. 2009). Results of numerous studies have shown that this bacterium may play various functions—it may be primary or secondary symbionts of blood- or plant sap-sucking insects (Thao et al. 2000; Russell et al. 2003; Trowbridge et al. 2006; Jousselin et al. 2013), plant pathogens vectored by planthoppers (Bressan 2014), or male killers in the parasitic wasp Nasonia vitripennis (Huger et al. 1985; Gherna et al. 1991). It has been also revealed that Arsenophonus may be horizontally and vertically transmitted to the next generation (Jousselin et al. 2013); however, the mode of transovarial transmission of this microorganism is poorly understood. To our knowledge, the occurrence of the bacterium Arsenophonus inside the cells of other bacterium has never been described in any insects. It is very difficult to interpret this phenomenon, but it may be assumed after von Dohlen et al. (2001) and Michalik et al. (2014) that, like the bacterium Sodalis in mealybugs and in the green leafhopper C. viridis, the bacterium Arsenophonus may be protected in the cytoplasm of other bacteria from the immune system of the host insect. This hypothesis is supported by observations that Arsenophonus may play various roles—from symbiotic to parasitic. Thus, its association with host insects may be more or less ancient. This, in turn, suggests that the more recently acquired bacterium Arsenophonus retained their virulence factors which might be detected by the insect immune system.
It should be stressed that Arsenophonus has not been found in the two species of Macrosteles collected in Japan: Macrosteles striifrons and M. sexnotatus (Ishii et al. 2013), but it has been detected in the species M. quadrilineatus collected in the USA (Bennett and Moran 2013). According to Bennett and Moran (2013), Arsenophonus does not represent an obligate symbiont of M. quadrilineatus. The distribution of Arsenophonus in the body of M. quadrilineatus as well as the mode of their transmission from mother to offspring remains unknown. The occurrence of Arsenophonus only in some species of Macrosteles and its absence in geographically isolated species strongly support the view of a recent association between these hemipterans and Arsenophonus. To verify this hypothesis, further molecular and ultrastructural studies on symbionts in other Macrosteles species are needed.
Earlier papers (Buchner 1925, 1965; Müller 1962) and more recent studies (Cheung and Purcell 1999; Sacchi et al. 2008; Michalik et al. 2014; this study) indicated that in Cicadomorpha, the symbiotic microorganisms migrate to the ovaries in a similar way, i.e., via the cytoplasm of follicular cells. Thus, in spite of the enormous diversity of types of symbionts living in Cicadomorpha, the mode of their transmission from mother to the offspring seems to be more uniform.
References
Baumann P (2005) Biology of bacteriocyte-associated endosymbionts of plant sup-sucking insects. Annu Rev Microbiol 59:155–189
Baumann P (2006) Diversity of prokaryote-insect associations within the Sternorrhyncha (psyllids, whiteflies, aphids, mealybugs). In: Miller TA, Bourtzis K (eds) Insect symbiosis, vol 2, Contemporary Topics in Entomology Series. CRC, Boca Raton, pp 1–24
Baumann L, Thao ML, Hess JM, Johnson MW, Baumann P (2002) The genetic properties of the primary endosymbionts of mealybugs differ from those of other endosymbionts of plant sap-sucking insects. Appl Environ Microbiol 68:3198–3205
Baumann P, Moran NA, Baumann L (2006) Bacteriocyte-associated endosymbionts of insects. Prokaryotes 1:403–438
Bennett GM, Moran NA (2013) Small, smaller, smallest: the origin and evolution of ancient dual symbioses in a phloem-feeding insect. Genome Biol Evol 5:1675–1688
Biliński S (1998) Introductory remarks. Folia Histochem Cytobiol 3:143–145
Brčak J (1979) Leafhopper and planthopper vectors of plant disease agents in central and southern Europe. In: Maramorosch K, Harris KF (eds) Leafhopper vectors and plant disease agents. Academic, New York, pp 97–154
Bressan A (2014) Emergence and evolution of Arsenophonus bacteria as insect-vectored plant pathogens. Infect Genet Evol 22:81–90
Bressan A, Arneodo J, Simonato M, Haines WP, Boudon-Padieu E (2009) Characterization and evolution of two bacteriome-inhabiting symbionts in cixiid planthoppers (Hemiptera: Fulgoromorpha: Pentastirini). Environ Microbiol 11:3265–3279
Buchner P (1925) Studien an intracellularen Symbionten. V. Die symbiotischen Einrichtungen der Zikaden. Z Morphol Ökol Tiere 4:88–245
Buchner P (1965) Endosymbiosis of animals with plant microorganisms. Interscience, New York
Büning J (1994) The ovary of Ectognatha, the insects s.str. In: Büning J (ed) The insect ovary: ultrastructure, previtellogenic growth and evolution. Chapman and Hall, London, pp 31–305
Cheung W, Purcell AH (1999) Invasion of bacteroids and BEV bacterium into oocytes of the leafhopper Euscelidius variegatus Kirschbaum (Homoptera: Cicadellidae): an electron microscopic study. Zool Stud 38:69–75
Douglas AE (1998) Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol 43:17–37
Douglas AE (2003) The nutritional physiology of aphids. Adv Insect Physiol 31:73–140
Douglas AE (2006) Phloem sap feeding by animals: problems and solutions. J Exp Bot 57:747–754
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214
Gatehouse LN, Sutherland P, Forgie SA, Kaji R, Christeller JT (2011) Molecular and histological characterization of primary (Betaproteobacteria) and secondary (Gammaproteobacteria) endosymbionts of three mealybug species. Appl Environ Microbiol 78:1187–1197
Gherna RL, Werren JH, Weisburg W, Cote R, Woese CR, Mandelco L et al (1991) Arsenophonus nasoniae gen. nov., sp. nov., the causative agent of the son-killer trait in the parasitic wasp Nasonia vitripennis. Int J Syst Bacteriol 41:563–565
Hall TA (1999) BIOEDIT: an user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Huger AM, Skinner SW, Werren JH (1985) Bacterial infections associated with the son-killer trait in the parasitoid wasp, Nasonia (= Mormoniella) vitripennis (Hymenoptera, Pteromalidae). J Invertebr Pathol 46:272–280
Ishii Y, Matsuura Y, Kakizawa S, Nikoh N, Fukatsu T (2013) Diversity of bacterial endosymbionts associated with Macrosteles leafhoppers vectoring phytopathogenic phytoplasmas. Appl Environ Microbiol 79:5013–5022
Jousselin E, Coeur d’Acier A, Vanlerberghe-Masutti F, Duron O (2013) Evolution and diversity of Arsenophonus endosymbionts in aphids. Mol Ecol 22:260–270
Koga R, Bennett GM, Cryan JR, Moran NA (2013) Evolutionary replacement of symbionts in an ancient and diverse insect lineage. Environ Microbiol 15:2073–2081
Kono M, Koga R, Shimada M, Fukatsu T (2008) Infection dynamics of coexisting beta- and gammaproteobacteria in the nested endosymbiotic system of mealybugs. Appl Environ Microbiol 74:4175–4184
Kuechler SM, Gibbs G, Burckhardt D, Dettner K, Hartung V (2013) Diversity of bacterial endosymbionts and bacteria-host co-evolution in Gondwanan relict moss bugs (Hemiptera: Coleorrhyncha: Peloridiidae). Environ Microbiol 15:2031–2042
Marzorati M, Alma A, Sacchi L, Pajoro M, Palermo S, Brusetti L et al (2006) A novel Bacteroidetes symbiont is localized in Scaphoideus titanus, the insect vector of flavescence dorée in Vitis vinifera. Appl Environ Microbiol 72:1467–1475
McCutcheon JP, McDonald BR, Moran NA (2009) Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci U S A 106:15394–15399
McCutcheon JP, Moran NA (2010) Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol Evol 2:708–718
McCutcheon JP, von Dohlen CD (2011) An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr Biol 21:1366–1372
Michalik A, Jankowska W, Kot M, Gołas A, Szklarzewicz T (2014) Symbiosis in the green leafhopper, Cicadella viridis (Hemiptera, Cicadellidae). Association in statu nascendi? Arthropod Struct Dev 43:579–587
Michalik A, Jankowska W, Szklarzewicz T (2009) Ultrastructure and transovarial transmission of endosymbiotic microorgansms in Conomelus anceps and Metcalfa pruinosa (Insecta, Hemiptera, Fulgoromorpha). Folia biol (Kraków) 57:131–137
Montlor CB, Maxmen A, Purcell AH (2002) Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol 27:189–195
Moran NA, Dale C (2006) Molecular interactions between bacterial symbionts and their hosts. Cell 126:453–465
Moran NA, Dale C, Dunbar H, Smith WA, Ochman H (2003) Intracellular symbionts of sharpshooters (Insecta: Hemiptera: Cicadellinae) form a distinct clade with a small genome. Environ Microbiol 5:116–126
Moran NA, Tran P, Gerardo NM (2005) Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol 71:8802–8810
Müller HJ (1962) Neuere Vorstellungen über Verbreitung und Phylogenie der Endosymbiosen der Zikaden. Z Morphol Ökol Tiere 51:190–210
Noda H (1977) Histological and histochemical observation of intracellular yeast-like symbiotes in the fat body of the small brown planthopper, Laodelphax striatellus (Homoptera: Delphacidae). Appl Entomol Zool 12:134–141
Noda H, Watanabe K, Kawai S, Yukuhiro F, Miyoshi T, Tomizawa M et al (2012) Bacteriome-associated endosymbionts of the green rice leafhopper Nephotettix cincticeps (Hemiptera: Cicadellidae). Appl Entomol Zool 47:217–225
Nováková E, Hypša V, Moran NA (2009) Arsenophonus, an emerging clade of intracellular symbionts with a broad host distribution. BMC Microbiol 9:143
Oliver KM, Russell JA, Moran NA, Hunter MS (2003) Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci U S A 100:1803–1807
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818
Rambaut A (2008) FigTree v. 1.4.0. Available http://tree.bio.ed.ac.uk/software/figtree/. Accessed 2013 May 20 Rambaut
Russell JA, Latorre A, Sabater-Muñoz B, Moya A, Moran NA (2003) Side-stepping secondary symbionts: widespread horizontal transfer across and beyond the Aphidoidea. Mol Ecol 12:1061–1075
Sacchi L, Genchi M, Clementi E, Bigliardi E, Avanzatti AM, Pajoroi M et al (2008) Multiple symbiosis in the leafhopper Scaphoideus titanus (Hemiptera: Cicadellidae): details of transovarial transmission of Cardinium sp. and yeast-like endosymbionts. Tissue Cell 40:231–242
Scarborough CL, Ferrari J, Godfray HCJ (2005) Aphid protected from pathogen by endosymbiont. Science 310:1781
Szklarzewicz T, Grzywacz B, Szwedo J, Michalik A (2015) Bacterial symbionts of the leafhopper Evacanthus interruptus (Linnaeus, 1758) (Insecta, Hemiptera, Cicadellidae: Evacanthinae). Protoplasma. doi:10.1007/s00709-015-0817-2
Takiya DM, Tran P, Dietrich CH, Moran NA (2006) Co-cladogenesis spanning three phyla: leafhoppers (Insecta: Hemiptera: Cicadellidae) and their dual bacterial symbionts. Mol Ecol 15:4175–4191
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thao ML, Clark MA, Baumann L, Brennan EB, Moran NA, Baumann P (2000) Secondary endosymbionts of psyllids have been acquired multiple times. Curr Microbiol 41:300–304
Thao ML, Gullan PJ, Baumann P (2002) Secondary (γ-proteobacteria) endosymbionts infect the primary (β-proteobacteria) endosymbionts of mealybugs multiple times and coevolve with their host. Appl Environ Microbiol 68:3190–3197
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Trowbridge RE, Dittmar K, Whiting MF (2006) Identification and phylogenetic analysis of Arsenophonus- and Photorhabdus-type bacteria from adult Hippoboscidae and Streblidae (Hippoboscoidea). J Invertebr Pathol 91:64–68
Urban J, Cryan J (2012) Two ancient bacterial endosymbionts have coevolved with the planthoppers (Insecta: Hemiptera: Fulgoroidea). BMC Evol Biol 12:87
von Dohlen CD, Kohler S, Alsop ST, McManus WR (2001) Mealybug β-proteobacterial endosymbionts contain γ-proteobacterial symbionts. Nature 412:433–435
Wangkeeree J, Miller TA, Hanboonsong Y (2011) Predominant bacteria symbionts in the leafhopper Matsumuratettix hiroglyphicus—the vector of sugarcane white leaf phytoplasma. Bull Insectol 64:215–216
Wu D, Daugherty SC, Van Aken SE, Pai GH, Watkins KL, Khouri H (2006) Metabolic complementarity and genomics of the dual symbiosis of sharpshooters. PLoS Biol 4, e188
Zahniser JN, Dietrich CH (2010) Phylogeny of the leafhopper subfamily Deltocephalinae (Hemiptera: Cicadellidae) based on molecular and morphological data with a revised family-group classification. Syst Entomol 35:489–511
Zahniser JN, Dietrich CH (2013) A review of the tribes of Deltocephalinae (Hemiptera: Auchenorrhyncha: Cicadellidae). Eur J Taxon 45:1–211
Acknowledgments
We are greatly indebted to M.Sc. Ada Jankowska and Dr. Olga Woźnicka for their skilled technical assistance. We would like to thank the editor and two anonymous reviewers for their constructive suggestions and comments which helped us to improve the manuscript. This work was supported by funds from research grant K/DSC/001743.
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Hanns H. Kassemeyer
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kobiałka, M., Michalik, A., Walczak, M. et al. Sulcia symbiont of the leafhopper Macrosteles laevis (Ribaut, 1927) (Insecta, Hemiptera, Cicadellidae: Deltocephalinae) harbors Arsenophonus bacteria. Protoplasma 253, 903–912 (2016). https://doi.org/10.1007/s00709-015-0854-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-015-0854-x