Abstract
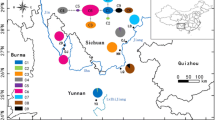
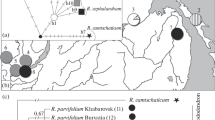
Based on three chloroplast markers (psbA-trnH, trnL-trnF, and trnT-trnL), we identified eight haplotypes in 23 samples of Litsea coreana collected from seven provinces across southern and southwestern China. The Hap6 was the most widely distributed haplotype. The genetic diversity was highest in Sichuan Province and its adjacent regions. Therefore, these regions should be areas of priority for conservation of L. coreana in the future. In addition, our molecular dating analyses indicated that the uplift of the Tibetan Plateau and past climate changes accounted for the diversification of L. coreana in China. Finally, we found that the chemical components of L. coreana were significantly affected by both collection time and postharvest handling methods.





Similar content being viewed by others
References
Avise JC (2000) Phylogeography: the history and formation of species. Harvard University Press, Cambridge
Avise JC (2001) Phylogeography. The history and formation of species. President and Fellows of Harvard College. United States of America
Bilton DT, Mirol PM, Mascheretti S, Fredga K, Zima J, Searle JB (1998) Mediterranean Europe as an area of endemism for small mammals rather than a source for northwards postglacial colonization. Proc Royal Soc B Biol Sci 265:1219–1226
Bustamante RO, Walkowiak A, Henriquez CA, Serey I (1996) Bird frugivory and the fate of seeds of Cryptocarya alba (Lauraceae) in the Chilean matorral. Revista Chilena de Historia Natural 69:357–363
Byrne M (2008) Evidence for multiple refugia at different time scales during Pleistocene climatic oscillations in southern Australia inferred from phylogeography. Quatern Sci Rev 27:2576–2585
Chanderbali AS, van der Werff H, Renner SS (2001) Phylogeny and historical biogeography of Lauraceae: evidence from the chloroplast and nuclear genomes. Ann Mo Bot Gard 88:104–134
Coulon A, Cosson JF, Angibault JM, Cargnelutti B, Galan M, Morellet N, Petit E, Aulagnier S, Hewison AJ (2004) Landscape connectivity influences gene flow in a roe deer population inhabiting a fragmented landscape: an individual-based approach. Mol Ecol 13:2841–2850
Datta A, Rawat GS (2008) Dispersal modes and spatial patterns of tree species in a tropical forest in Arunachal Pradesh, northeast India. Trop Conserv Sci 1:163–185
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214
Felsenstein J (1985) Confidence-limits on phylogenies—an approach using the bootstrap. Evolution 39:783–791
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hewitt GM (1999) Post-glacial re-colonization of European biota. Biol J Linn Soc 68:87–112
Hewitt G (2000) The genetic legacy of the Quaternary ice ages. Nature 405:907–913
Hewitt GM (2004) Genetic consequences of climatic oscillations in the Quaternary. Philos Trans Royal Soc London Ser B Biol Sci 359:183–195 discussion 195
Kaneko Y, Lian C, Watanabe S, Shimatani K, Sakio H, Noma N (2012) Development of microsatellites in Machilus thunbergii (Lauraceae), a warm-temperate coastal tree species in Japan. Am J Bot 99:e265–e267
Lenormand T (2002) Gene flow and the limits to natural selection. Trends Ecol Evol 17:183–189
Li JJ, Wen SX, Zhang QS, Wang FB, Zheng BX, Li BY (1979) A discussion on the period, amplitude and type of the uplift of the Qinghai–Xizang Plateau. Sci China Mathe 22:1314–1328
Li S, Pearl DK, Doss H (2000) Phylogenetic tree construction using Markov Chain Montecarlo. J Am Stat Assoc 95:493–508
Li JZ, Qiu J, Liao WB, Jin JH (2009) Eocene fossil Alseodaphne from Hainan Island of China and its paleoclimatic implications. Sci China Ser D Earth Sci 52:1537–1542
Long Y, Wan H, Yan F, Xu C, Lei G, Li S, Wang R (2006) Glacial effects on sequence divergence of mitochondrial COII of Polyura eudamippus (Lepidoptera: Nymphalidae) in China. Biochem Genet 44:361–377
Parrish J (1993) The paleogeography of the opening South Atlantic. In: George W, Lavocat R (eds) The Africa-South America connection. Clar-endon Press, Oxford, pp 8–27
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256
Rambaut A, Charleston M (2001) TreeEdit Ver 1.0. University of Oxford, Oxford
Rambaut A, Drummond AJ (2007) Tracer v1.4. http://beast.bio.ed.ac.uk/Tracer
Rohwer JG (2000) Toward a phylogenetic classification of the Lauraceae: evidence from matK Sequences. Syst Bot 25(1):60–71
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Royden LH, Burchfiel BC, van der Hilst RD (2008) The geological evolution of the Tibetan Plateau. Science 321:1054–1058
Sang T, Crawford D, Stuessy T (1997) Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am J Bot 84:1120
Sclater J, Hellinger S, Trapscott C (1977) The paleobathymetry of the Atlantic Ocean from the Jurassic to the present. J Geol 85:509–952
Shi YF, Li JJ, Li BY (1998) Uplift and environmental changes of Qinghai-Tibetan Plateau in the Late Cenozoic. Guangdong Science and Technology Press, Guangzhou
Slatkin M (1987) Gene flow and the geographic structure of natural populations. Science 236:787–792
Song J, Li B (1746) Jianwei County Annals, vol 1
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22(21):2688–2690
Swofford DL (2000) PAUP*. Phylogenetic analysis using parsimony (* and other methods). Version 4. Sinauer Associates, Sunderland Massachusetts
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17:1105–1109
Tate JA, Simpson BB (2003) Paraphyly of Tarasa (Malvaceae) and diverse origins of the polyploid species. Syst Bot 28:723–737
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Wang EC, Fan C, Wang G, Shi XH, Chen LZ, Chen ZK (2006) Deformational and geomorphic processes in the Ailao Shan-Diancang Range, West Yunnan. Quat Sci 26:220–227
Wang L, Abbott RJ, Zheng W, Chen P, Wang Y, Liu J (2009) History and evolution of alpine plants endemic to the Qinghai-Tibetan Plateau: Aconitum gymnandrum (Ranunculaceae). Mol Ecol 18:709–721
Webb T, Bartlein P (1992) Global changes during the last 3 million years: climatic controls and biotic responses. Annu Rev Ecol Syst 23:141–173
Acknowledgments
This research was supported by the National Natural Science Foundation of P. R. China (No. 81072995, No. 81274188). The authors are very grateful to Dr. Jin-feng Chen (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for his valuable scripts to phylogeny construction.
Author information
Authors and Affiliations
Corresponding author
Additional information
F. Han and L. Xu contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
606_2014_1031_MOESM1_ESM.doc
Supplementary material 1 (DOC 91 kb) Supplementary Doc 1 BI tree based on three chloroplast markers psbA-trnH, trnL-trnF, and trnT-trnL, with eight sequences from Monimiaceae, Hernandiaceae, and Gomortegaceae as outgroups
606_2014_1031_MOESM2_ESM.pdf
Supplementary material 2 (PDF 11 kb) GenBank accession numbers for the psbA-trnH, trnL-trnF, and trnT-trnL markers from Lauraceae
Rights and permissions
About this article
Cite this article
Han, F., Xu, L., Peng, Y. et al. The pattern of genetic diversity within Litsea coreana (Lauraceae) in China: an implication for conservation. Plant Syst Evol 300, 2229–2238 (2014). https://doi.org/10.1007/s00606-014-1031-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-014-1031-y