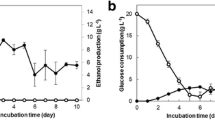
Abstract
Xylose is one of the most abundant lignocellulosic components, but it cannot be used by R. oryzae for fumaric acid production. Here, we applied high-throughput RNA sequencing to generate two transcriptional maps of R. oryzae following fermentation in glucose or xylose. The differential expression analysis showed that, genes involved in amino acid metabolism, fatty acid metabolism, and gluconeogenesis, were up-regulated in response to xylose. Moreover, we discovered the potential presence of oxidative stress in R. oryzae during xylose fermentation. To adapt to this unfavorable condition, R. oryzae displayed reduced growth and induce of a number of antioxidant enzymes, including genes involved in glutathione, trehalose synthesis, and the proteasomal pathway. These responses might divert the flow of carbon required for the accumulation of fumaric acid. Furthermore, using high-throughput RNA sequencing, we identified a large number of novel transcripts and a substantial number of genes that underwent alternative splicing. Our analysis provides remarkable insight into the mechanisms underlying xylose fermentation by R. oryzae. These results may reveal potential target genes or strategies to improve xylose fermentation.









Similar content being viewed by others
References
Tai C, Keshwani DR (2014) Enzyme adsorption and cellulose conversion during hydrolysis of dilute-acid-pretreated corn stover. Energy Fuels 28:1956–1961
Ji XJ, Nie ZK, Huang H, Ren LJ, Peng C, Ouyang PK (2011) Elimination of carbon catabolite repression in Klebsiella oxytoca for efficient 2, 3-butanediol production from glucose–xylose mixtures. Appl Microbiol Biotechnol 89:1119–1125
Zhang K, Yu C, Yang ST (2015) Effects of soybean meal hydrolysate as the nitrogen source on seed culture morphology and fumaric acid production by Rhizopus oryzae. Process Biochem 50:173–179
Xu Q, Li S, Huang H, Wen J (2012) Key technologies for the industrial production of fumaric acid by fermentation. Biotechnol Adv 30:1685–1696
Wang G, Huang D, Li Y, Wen J, Jia X (2015) A metabolic-based approach to improve xylose utilization for fumaric acid production from acid pretreated wheat bran by Rhizopus oryzae. Bioresour Technol 180:119–127
Wen S, Liu L, Nie KL, Deng L, Tan TW, Wang F (2013) Enhanced fumaric acid production by fermentation of xylose using a modified strain of Rhizopus arrhizus. BioResources 8:2186–2194
Liang S, Wang B, Pan L, Ye Y, He M, Han S, Zheng S, Wang X, Lin Y (2012) Comprehensive structural annotation of Pichia pastoris transcriptome and the response to various carbon sources using deep paired-end RNA sequencing. BMC Genom 13:738
Li J, Lin L, Li H, Tian C, Ma Y (2014) Transcriptional comparison of the filamentous fungus Neurospora crassa growing on three major monosaccharides d-glucose, d-xylose and l-arabinose. Biotechnol Biofuels 7:31
Wang J, Chen L, Huang S, Liu J, Ren X, Tian X, Qiao J, Zhang W (2012) RNA-seq based identification and mutant validation of gene targets related to ethanol resistance in cyanobacterial Synechocystis sp. PCC 6803. Biotechnol Biofuels 5:89
Yuan T, Ren Y, Meng K, Feng Y, Yang P, Wang S, Shi P, Wang L, Xie D, Yao B (2011) RNA-Seq of the xylose-fermenting yeast Scheffersomyces stipitis cultivated in glucose or xylose. Appl Microbiol Biotechnol 92:1237–1249
Crawford JE, Guelbeogo WM, Sanou A, Traoré A, Vernick KD, Sagnon N, Lazzaro BP (2010) De novo transcriptome sequencing in Anopheles funestus using Illumina RNA-seq technology. PLoS One 5:e14202
Abe A, Oda Y, Asano K, Sone T (2007) Rhizopus delemar is the proper name for Rhizopus oryzae fumaric-malic acid producers. Mycologia 99:714–722
Xu Q, Li S, Fu Y, Tai C, Huang H (2010) Two-stage utilization of corn straw by Rhizopus oryzae for fumaric acid production. Bioresour Technol 101:6262–6264
Yan L, Zhang H, Chen J, Lin Z, Jin Q, Jia H, Huang H (2009) Dilute sulfuric acid cycle spray flow-through pretreatment of corn stover for enhancement of sugar recovery. Bioresour Technol 100:1803–1808
Hipler U, Wollina U, Denning D, Hipler B (2000) Fluorescence analysis of reactive oxygen species (ROS) generated by six isolates of aspergillus fumigatus. Bioluminescence and Chemiluminescence, pp 411–414
Jiang L, Cai J, Wang J, Liang S, Xu Z, Yang ST (2010) Phosphoenolpyruvate-dependent phosphorylation of sucrose by Clostridium tyrobutyricum ZJU 8235: evidence for the phosphotransferase transport system. Bioresour Technol 101:304–309
Matsushika A, Nagashima A, Goshima T, Hoshino T (2013) Fermentation of xylose causes inefficient metabolic state due to carbon/energy starvation and reduced glycolytic flux in recombinant industrial Saccharomyces cerevisiae. PLoS One 8:e69005
Nitsche BM, Jørgensen TR, Akeroyd M, Meyer V, Ram AF (2012) The carbon starvation response of Aspergillus niger during submerged cultivation: insights from the transcriptome and secretome. BMC Genom 13:380
Novodvorska M, Hayer K, Pullan ST, Wilson R, Blythe MJ, Stam H, Stratford M, Archer DB (2013) Transcriptional landscape of Aspergillus niger at breaking of conidial dormancy revealed by RNA-sequencing. BMC Genom 14:246
Jozefczuk S, Klie S, Catchpole G, Szymanski J, Cuadros-Inostroza A, Steinhauser D, Selbig J, Willmitzer L (2010) Metabolomic and transcriptomic stress response of Escherichia coli. Mole Sys Biol 6(1):364
Rodríguez-Vargas S, Sánchez-García A, Martínez-Rivas JM, Prieto JA, Randez-Gil F (2007) Fluidization of membrane lipids enhances the tolerance of Saccharomyces cerevisiae to freezing and salt stress. Appl Environ Microbiol 73:110–116
Liu Y, Lv C, Xu Q, Li S, Huang H, Ouyang P (2015) Enhanced acid tolerance of Rhizopus oryzae during fumaric acid production. Bioprocess Biosyst Eng 38:323–328
Li Q, McNeil B, Harvey LM (2008) Adaptive response to oxidative stress in the filamentous fungus Aspergillus niger B1-D. Free Radic Biol Med 44:394–402
Kirsch M, Grooth De (2001) NAD(P)H, a directly operating antioxidant? FASEB J 15:1569–1574
Wang X, Jin M, Balan V, Jones AD, Li X, Li BZ, Dale BE, Yuan YJ (2014) Comparative metabolic profiling revealed limitations in xylose-fermenting yeast during co-fermentation of glucose and xylose in the presence of inhibitors. Biotechnol Bioeng 111:152–164
Li Q, Harvey LM, McNeil B (2009) Oxidative stress in industrial fungi. Crit Rev Biotechnol 29:199–213
Ye Y, Zhang L, Hao F, Zhang J, Wang Y, Tang H (2012) Global metabolomic responses of Escherichia coli to heat stress. J Proteome Res 11:2559–2566
Acknowledgments
This work was financially supported by the National Basic Research Program of China (No. 2013CB733605), National Science Foundation for Distinguished Young Scholars of China (No. 21225626), the National High Technology Research and Development Program of China (No. 2011AA02A206 and No. 2012AA022304), the National Natural Science Foundation of China (No. 21106065), and the Excellent Doctoral Dissertation Foundation of Guangdong Province (sybzzxm201217).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Xu, Q., Liu, Y., Li, S. et al. Transcriptome analysis of Rhizopus oryzae in response to xylose during fumaric acid production. Bioprocess Biosyst Eng 39, 1267–1280 (2016). https://doi.org/10.1007/s00449-016-1605-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-016-1605-x