Abstract
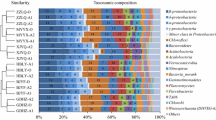
Full-scale applications of autotrophic nitrogen removal technologies for the treatment of digested sludge liquor have proliferated during the last decade. Among these technologies, the aerobic/anoxic deammonification process (DEMON) is one of the major applied processes. This technology achieves nitrogen removal from wastewater through anammox metabolism inside a single bioreactor due to alternating cycles of aeration. To date, microbial community composition of full-scale DEMON bioreactors have never been reported. In this study, bacterial community structure of a full-scale DEMON bioreactor located at the Apeldoorn wastewater treatment plant was analyzed using pyrosequencing. This technique provided a higher-resolution study of the bacterial assemblage of the system compared to other techniques used in lab-scale DEMON bioreactors. Results showed that the DEMON bioreactor was a complex ecosystem where ammonium oxidizing bacteria, anammox bacteria and many other bacterial phylotypes coexist. The potential ecological role of all phylotypes found was discussed. Thus, metagenomic analysis through pyrosequencing offered new perspectives over the functioning of the DEMON bioreactor by exhaustive identification of microorganisms, which play a key role in the performance of bioreactors. In this way, pyrosequencing has been proven as a helpful tool for the in-depth investigation of the functioning of bioreactors at microbiological scale.





Similar content being viewed by others
References
Rodriguez-Sanchez A, Gonzalez-Martinez A, Martinez-Toledo MV, Garcia-Ruiz MJ, Osorio F, Gonzalez-Lopez J (2014) The effect of influent characteristics and operational conditions over the performance and microbial community structure of partial nitritation reactors. Water 6:1905–1924
Kotay SM, Mansell BL, Hogsett M, Pei H, Goel R (2013) Anaerobic ammonia oxidation (ANAMMOX) for side-stream treatment of anaerobic digester filtrate process performance and microbiology. Biotechnol Bioeng 110:1180–1192
Shen LD, Liu S, Lou LP, Liu WP, Xu XY, Zheng P, Hu BI (2013) Broad distribution of diverse anaerobic ammonium-oxidizing bacteria in chinese agricultural soils. Appl Environ Microbiol 79:6167–6172
Ganesh S, Parris DJ, DeLong EF, Stewart FJ (2014) Metagenomic analysis of size-fractionated picoplankton in a marine oxygen minimum zone. ISMEJ 8(1):187–211
Thamdrup B, Jensen MM, Ulloa O, Faria L, Escribano R (2006) Anaerobic ammonium oxidation in the oxygen-deficient waters off northern Chile. Limnol Oceanogr 51(5):2145–2156
Innerebner G, Insam H, Franke-Whittle IH, Wett B (2007) Identification of anammox bacteria in a full-scale deammonification plant making use of anaerobic ammonia oxidation. Syst Appl Microbiol 30(5):408–412
Gonzalez-Martinez A, Rodriguez-Sanchez A, Martinez-Toledo MV, Garcia-Ruiz MJ, Hontoria E, Osorio-Robles F, Gonzalez-Lopez J (2014) Effect of ciprofloxacin antibiotic on the partial-nitritation process and bacterial community structure of a submerged biofilter. Sci Total Environ 476–477:276–287
Dalahmeh SS, Jönson H, Hylander LD, Hui N, Yu D, Pell M (2014) Dynamics and functions of bacterial communities in bark, charcoal and sand filters treating greywater. Water Res 54:21–32
Han D, Kang I, Ha HK, Kim HC, Kim OS, Lee BY, Cho JC, Hur HG, Lee YH (2014) Bacterial communities of surface mixed layer in the Pacific sector of the Western Arctic Ocean during sea-ice melting. PLoS One 9(1):e86887
Huang Q, Jiang H, Briggs BR, Wang S, Hou W, Li G, Wu G, Solis R, Arcilla CA, Abrajano T, Dong H (2013) Archaeal and bacterial diversity in acidic circumneutral hot springs in Philippines. FEMS Microbiol 85:452–464
Livermore JA, Jin YO, Arnseth RW, LePuil M, Mattes TE (2013) Microbial community dynamics during acetate biostimulation of RDX-contaminated groundwater. Environ Sci Technol 47:7672–7678
Zekker I, Rikmann E, Tenno T, Saluste A, Tomingas M, Menert A, Loorits L, Lemmiksoo V, Tenno T (2012) Achieving nitritation and anammox enrichment in a single moving-bed biofilm reactor treating reject water. Environ Technol 33(6):703–710
Mozumder MSI, Picioreanu C, van Loosdrecht MCM, Volcke EIP (2014) Effect of heterotrophic growth on autotrophic nitrogen removal in a granular sludge reactor. Environ Technol 35(8):1027–1037
Sorokin DY, Lücker S, Vejmelkova D, Kostrikina NA, Kleerebezem R, Rijpstra WIC, Damsté JSS, Le Paslier D, Muyzer G, Wagner M, van Loosdrecht MCM, Daims H (2012) Nitrification expanded: discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISMEJ 6:2245–2256
Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, Wolcott RD (2008) Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol 8:43
Nakayama J (2010) Pyrosequence-based 16S rRNA profiling of gastro-intestinal microbiota. Biosci Microflora 29(2):83–96
Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ (2006) Microbial diversity in the deep sea and the underexplored “rare biosphere”. PNAS 103(32):12115–12120
Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG, Edrington TS (2008) Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol 8:125
R Development Core Team (2008) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Park H, Rosenthal A, Ramalingam K, Fillos J, Chandran K (2010) Linking community profiles, gene expression and N-removal in anammox bioreactors treating municipal anaerobic digestion reject water. Environ Sci Technol 44(16):6110–6116
Kant R, van Passel MWJ, Sangwan P, Palva A, Lucas S, Copeland A et al (2011) Genome sequence of “Pedosphaera parvula” Ellin514, an aerobic Verrucomicrobial isolate from pasture soil. J Bacteriol 193:2900–2901
Sangwan P, Kovac S, Davis KER, Sait M, Janssen PH (2005) Detection and Cultivation of Soil Verrucomicrobia. Appl Environ Microbiol 71(12):8402–8410
Galperin MY (2008) New feel for new phyla. Environ Microbiol 10:1927–1933
Chouari R, Le Paslier D, Dauga C, Daegelen P, Weissenbach J, Sghir A (2005) Novel major bacteria candidate division within a municipal anaerobic sludge digester. Appl Environ Microbiol 71(4):2145–2153
Haldar S, Choudhury SR, Sengupta S (2011) Genetic and functional diversities of bacterial communities in the rhizosphere of Arachis hypogaea. Antonie Van Leeuwenhoek 100:161–170
Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T et al (2002) Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc Natl Acad Sci USA 99:996–1001
Bruggemann H, Baumer S, Fricke WF, Wiezer A, Liesegang H, Decker I et al (2003) The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc Natl Acad Sci USA 100:1316–1321
Nölling J, Breton G, Omelchenko MV, Kira S, Zeng Q, Gibson R et al (2001) Genome sequence and comparative analysis of the solvent-producing Bacterium Clostridium acetobutylicum. J Bacteriol 183(16):4823–4838
Miura Y, Watanabe Y, Okabe S (2007) Significance of Chloroflexi in performance of submerged membrane bioreactors (MBR) treating municipal wastewater. Environ Sci Technol 41(22):7787–7794
Nelson MC, Morrison M, Yu Z (2011) A meta-analysis of the microbial diversity observed in anaerobic digesters. Biores Technol 102:3730–3739
Strous M, Fuerst JA, Kramer EHM, Logemann S et al (1999) Missing lithotroph identified as new planctomycete. Nature 400:446–449
Yoon JH, Kim IG, Oh TK, Park YH (2004) Microbulbifer maritimus sp. nov., isolated from an intertidal sediment from the Yellow Sea, Korea. Int J Syst Evol Microbiol 54(4):1111–1116
Miyazaki M, Nogi Y, Ohta Y, Hatada Y, Fujiwara Y, Ito S, Horikoshi K (2008) Microbulbifer agarilyticus sp. nov. and Microbulbifer thermotolerans sp. nov., agar-degrading bacteria isolated from deep-sea sediment. Int J Syst Evol Microbiol 58(5):1128–1133
Gonzalez JM, Mayer F, Moran MA, Hodson RE, Whitman WB (1997) Microbulbifer hydrolyticus gen. nov., sp. nov., and Marinobacterium georgiense gen. nov., sp. nov., two marine bacteria from a lignin-rich pulp mill waste enrichment community. Int J Syst Bacteriol 47:369–376
Guo F, Zhang S-H, Yu X, Wei B (2011) Variations of both bacterial community and extracellular polymers: the inducements of increase of cell hydrophobicity from biofloc to aerobic granule sludge. Biores Technol 102(11):6421–6428
Gonzalez-Martinez A, Calderon K, Albuquerque A, Hontorio E, Gonzalez-Lopez J, Guisado IM, Osorio F (2013) Biological and technical study of a partial-SHARON reactor at laboratory scale: effect of hydraulic retention time. Bioprocess Biosyst Eng 36:173–184
Yoo SH, Weon HY, Jang HB, Kim BY, Kwon SW, Go SJ, Stackebrandt E (2007) Sphingobacterium composti sp. nov., isolated from cotton-waste composts. Int J Syst Evol Microbiol 57(7):1590–1593
Bao HX, Ma XP, Wang J, Jing K, Shen ML, Wang AJ (2012) Performance of denitrifying phosphorous removal process employing nitrite as electron acceptor and the characterization of dominant functional populations. Adv Mater Res 455–456:1019–1024
Nagashima S, Kamimura A, Shimizu T, Nakamura-Isaki S, Aono E, Sakamoto K et al (2012) Complete genome sequence of phototrophic betaproteobacterium Rubrivivax gelatinosus IL144. J Bacteriol 194(13):3541–3542
Xie B, Lv Z, Hu C, Yang X, Li X (2013) Nitrogen removal through different pathways in an aged refuse bioreactor treating mature landfill leachate. Appl Microbiol Biotechnol 97(20):9225–9234
Lam P, Jensen MM, Lavik G, McGinnis DF, Müller B, Schubert CJ, Amann R, Thamdrup B, Kuypers MMM (2007) Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc Natl Acad Sci USA 104(17):7104–7109
Fahrbach M, Kuever J, Remesch M, Huber BE, Kämpfer P, Dott W, Hollender J (2008) Steroidobacter denitrificans gen. nov., sp. nov., a steroidal hormone-degrading gammaproteobacterium. Int J Syst Evol Microbiol 58(Pt 9):2215–2223
Glavina Del Rio T, Abt B, Spring S, Lapidus A, Nolan M, Tice H et al (2010) Complete genome sequence of Chitinophaga pinensis type strain (UQM 2034). Stand Gen Sci 2(1):87–95
Chen HJ, Lin YZ, Fanjiang JM, Fan C (2013) Microbial community and treatment ability investigation in AOAO process for the optoelectronic wastewater treatment using PCR-DGGE biotechnology. Biodegradation 24(2):227–243
Suen G, Weimer PJ, Stevenson DM, Aylward FO, Boyum J, Deneke J et al (2011) The complete genome sequence of Fibrobacter succinogenes S85 reveals a cellulolytic and metabolic specialist. PloS One 6(4):e18814
Woyke T, Chertkov O, Lapidus A, Nolan M, Lucas S, Del Rio TG et al (2011) Complete genome sequence of the gliding freshwater bacterium Fluviicola taffensis type strain (RW262). Stand Gen Sci 5(1):21–29
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gonzalez-Martinez, A., Rodriguez-Sanchez, A., Muñoz-Palazon, B. et al. Microbial community analysis of a full-scale DEMON bioreactor. Bioprocess Biosyst Eng 38, 499–508 (2015). https://doi.org/10.1007/s00449-014-1289-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-014-1289-z