Abstract
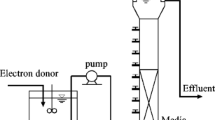
A pilot-scale reactor treating domestic sewage was operated to promote anaerobic digestion and denitrification using endogenous electron donors. While 55 % of organic matter was removed, nitrogen and sulfur showed a different dynamics during the operation. Pyrosequencing analysis clarified this behavior revealing that specific microbial communities inhabited the anaerobic (47.05 % of OTUs) and anoxic (31.39 % of OTUs) chambers. Analysis of 16S rRNA gene partial sequences obtained through pyrosequencing revealed a total of 1727 OTUs clustered at a 3 % distance cutoff. In the anaerobic chamber, microbial community was comprised of fermentative, syntrophic and sulfate-reducing bacteria. The majority of sequences were related to Aminobacterium and Syntrophorhabdus. In the anoxic chamber, the majority of sequences were related to mixotrophic and strictly autotrophic denitrifiers Arcobacter and Sulfuricurvum, respectively, both involved in sulfur-driven denitrification. These results show that pyrosequencing was a powerful tool to investigate the microbial panorama of a complex system, providing new insights to the improvement of the system.





Similar content being viewed by others
References
Foresti E, Zaiat M, Vallero M (2006) Anaerobic processes as the core technology for sustainable domestic wastewater treatment: consolidated applications, new trends, perspectives, and challenges. Rev Environ Sci Biotechnol 5:3–19
von Sperling M (2015) Comparison of simple, small, full-scale sewage treatment systems in Brazil: UASB–maturation ponds–coarse filter; UASB–horizontal subsurface-flow wetland; vertical-flow wetland (first stage of French system). Water Sci Technol 71(3):329–337
Moraes BS, Orrú JG, Foresti E (2013) Nitrogen and sulfide removal from effluent of UASB reactor in a sequencing fed-batch biofilm reactor under intermittent aeration. J Biotechnol 164(3):378–385
Gonzalez-Martinez A, Osorio F, Rodriguez-Sanchez A, Martinez-Toledo MV, Gonzalez-Lopez J, Lotti T, van Loosdrecht MC (2015) Bacterial community structure of a lab-scale anammox membrane bioreactor. Biotechnol Prog 1:186–193
Sanz JL, Kochling T (2007) Molecular biology techniques used in wastewater treatment: an overview. Process Biochem 42:119–133
Glenn TC (2011) Field guide to next generation DNA sequencers. Mol Ecol Resour 15:759–769
Hu M, Wang X, Wen X, Xia Y (2012) Microbial community structures in different wastewater treatment plants as revealed by 454-pyrosequencing analysis. Bioresour Technol 117:72–79
Zhang T, Shao MF, Ye L (2012) 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J 6:1137–1147
Park S, Yu J, Byun I, Cho S, Park T, Lee T (2011) Microbial community structure and dynamics in a mixotrophic nitrogen removal process using recycled spent caustic under different loading conditions. Bioresour Technol 102:7265–7271
American Public Health Association/American Water Works Association/Water Environment Federation (APHA/AWWA/WEF) (2005) Standard methods for the examination of water and wastewater, 21st edn. Washington DC, USA
Griffiths RI, Whiteley AS, O’Donnell AG, Bailey MJ (2000) Rapid method for coextraction of dna and rna from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microbiol 66(12):5488–5491
Sul WJ, Cole JR, Jesus EC, Wang Q, Farris RJ, Fish JA, Tiedje JM (2011) Bacterial community comparisons by taxonomy-supervised analysis independent of sequence alignment and clustering. Proc Natl Acad Sci USA 35:14637–14642
Cardenas E, Wei-Min WU, Leigh ML, Carley J, Carroll S, Gentry T, Luo J, Watson D, Gu B, Ginder-Vogel M, Kitanidis PK, Jardine PM, Zhou J, Criddle CS, Marsh TL, Tiedje JM (2010) Significant association between sulfate-reducing bacteria and uranium-reducing microbial communities as revealed by a combined massively parallel sequencing-indicator species approach. Appl Environ Microbiol 76(20):6778–6786
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, Human Microbiome Consortium, Petrosino JF, Knight R, Birren BW (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J, Glöckner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acid Res 35:7188–7196
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71(3):1501–1506
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) A naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Lemos LN, Fulthorpe RR, Triplett EW, Roesch LFW (2011) Rethinking microbial diversity analysis in the high throughput sequencing era. J Microbiol Meth 86:42–51
Campbell BJ, Engel AS, Porter ML, Takai K (2006) The versatile ε-proteobacteria: keyplayers in sulphidic habitats. Nat Rev Microbiol 4:458–468
Collado L, Levican A, Perez J, Figueras MJ (2011) Arcobacterdefluvii sp. nov., isolated from sewage samples. Int J Syst Evol Microbiol 61(9):2155–2161
Kodama Y, Watanabe K (2004) Sulfuricurvum kujiense gen. nov., sp. nov., a facultatively anaerobic, chemolithoautotrophic, sulfur-oxidizing bacterium isolated from an underground crude-oil storage cavity. Int J Syst Evol Microbiol 54(6):2297–2300
De Gusseme B, De Schryver P, De Cooman M, Verbeken K, Boeckx P, Verstraete W, Boon N (2009) Nitrate-reducing, sulfide-oxidizing bacteria as microbial oxidants for rapid biological sulfide removal. FEMS Microbiol Ecol 67(1):151–161
McLellan SL, Huse SM, Mueller-Spitz SR, Andreishcheva EN, Sogin ML (2010) Diversity and population structure of sewage derived microorganisms in wastewater treatment plant influent. Environ Microbiol 12(2):378–392
Mahmood Q, Zheng P, Hu B, Jilani G, Azim MR, Wu D, Liu D (2009) Isolation and characterization of Pseudomonas stutzeri QZ1 from an anoxic sulfide-oxidizing bioreactor. Anaerobe 15(4):108–115
Wen Y, Ren Y, Wei C-H, Li K-Y, Lin F-M, Chen X-Y (2010) A study on nitrogen removal efficiency of Pseudomonas stutzeri strains isolated from an anaerobic/anoxic/oxic wastewater treatment process. Afr J Biotechnol 9(6):869–873
Martin-Carnahan A, Joseph SW (2005) Genus I. Aeromonas Stanier 1943, 213AL. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM (eds) Bergey’s manual of systematic bacteriology, 2nd edn. vol 2 (The Proteobacteria), part B (The Gammaproteobacteria), Springer, New York, pp 557–558
Lim YW, Lee SA, Kim SB, Yong HY, Yeon SH, Park YK, Jeong DW, Park JS (2005) Diversity of denitrifying bacteria isolated from Daejeon sewage treatment plant. J Microbiol 43(5):383–390
Fischer-Romero C, Tindall BJ, Jüttner F (1996) Tolumonasauensis gen. nov., sp. nov., a toluene-producing bacterium from anoxic sediments of a freshwater lake. Int J Syst Bacteriol 46(1):183–188
Dannenberg S, Kroder M, Dilling W, Cypionka H (1992) Oxidation of H2, organic compounds and inorganic sulfur compounds coupled to reduction of O2 or nitrate by sulfate-reducing bacteria. Arch Microbiol 158(2):93–99
Bergmann GT, Bates ST, Eilers KG, Lauber CL, Caporaso JG, Walters WA, Knight R, Fierer N (2011) The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol Biochem 43(7):1450–1455
Chouari R, Le Paslier D, Dauga C, Daegelen P, Weissenbach J, Sghir A (2005) Novel major bacteria candidate division within a municipal anaerobic sludge digester. Appl Environ Microbiol 71(4):2145–2153
Gonzalez-Martinez A, Rodriguez-Sanchez A, Muñoz-Palazon B, Garcia-Ruiz MJ, Osorio F, van Loosdrecht MC, Gonzalez-Lopez J (2015) Microbial community analysis of a full-scale DEMON bioreactor. Bioprocess Biosyst Eng 38(3):499–508
Liesack W, Bak F, Kreft JU, Stackebrandt E (1994) Holophaga foetida gen. nov., sp. nov., a new, homoacetogenic bacterium degrading methoxylated aromatic compounds. Arch Microbiol 162(1–2):85–90
Vartoukian SR, Palmer RM, Wade WG (2007) The division “Synergistes”. Anaerobe 13(3–4):99–106
Ramsay IR, Pullammanappallil PC (2001) Protein degradation during anaerobic wastewater treatment: derivation of stoichiometry. Biodegradation 12(4):247–257
Qiu YL, Sekiguchi Imachi H, Kamagata Y, Tseng IC, Cheng SS, Ohashi A, Harada H (2004) Identification and isolation of anaerobic, syntrophic phthalate isomer-degrading microbes from methanogenic sludges treating wastewater from terephthalate manufacturing. Appl Environ Microbiol 70:1617–1626
Hofmann K, Hammer E (1999) Anaerobic formation and degradation of toxic aromatic compounds in agricultural and communal sewage deposits. Chemosphere 38(11):2561–2568
Ariesyady HD, Ito T, Yoshiguchi K, Okabe S (2007) Phylogenetic and functional diversity of propionate-oxidizing bacteria in an anaerobic digester sludge. Appl Microbiol Biotechnol 75(3):673–683
Yamada T, Imachi H, Ohashi A, Harada H, Hanada S, Kamagata Y, Sekiguchi Y (2007) Bellilinea caldifistulae gen. nov., sp. nov. and Longilinea arvoryzae gen. nov., sp. nov., strictly anaerobic, filamentous bacteria of the phylum Chloroflexi isolated from methanogenic propionate-degrading consortia. Int J Syst Evol Microbiol 57(10):2299–2306
Yamada T, Sekiguchi Y, Hanada S, Imachi H, Ohashi A, Harada H, Kamagata Y (2006) Anaerolinea thermolimosa sp. nov., Levilinea saccharolytica gen. nov., sp. nov. and Leptolinea tardivitalis gen. nov., sp. nov., novel filamentous anaerobes, and description of the new clases Anaerolineae classis nov. And Caldilineae classis nov. In the bacterial phylum Chloroflexi. Int J Syst Evol Microbiol 56(6):1331–1340
Yamada T, Sekiguchi Y, Imachi H, Kamagata Y, Ohashi A, Harada H (2005) Diversity, localization, and physiological properties of filamentous microbes belonging to Chloroflexi subphylum I in mesophilic and thermophilic methanogenic sludge granules. Appl Environ Microbiol 71(11):7493–7503
Acknowledgments
This work was funded by FAPESP - Fundação de Amparo à Pesquisa do Estado de São Paulo (Process numbers: 2007/58659-7, 2010/03286-4 and 2010/01735-6) and by CNPq - Conselho Nacional de Desenvolvimento Científico e Tecnológico.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Saia, F.T., Souza, T.S.O., Duarte, R.T.D. et al. Microbial community in a pilot-scale bioreactor promoting anaerobic digestion and sulfur-driven denitrification for domestic sewage treatment. Bioprocess Biosyst Eng 39, 341–352 (2016). https://doi.org/10.1007/s00449-015-1520-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-015-1520-6