Abstract
Acute respiratory distress syndrome (ARDS) is today a leading cause of hospitalization in intensive care unit (ICU). ARDS and pneumonia are closely related to critically ill patients; however, the etiologic agent is not always identified. The presence of human herpes simplex virus 1, human cytomegalovirus and Epstein–Barr virus in respiratory samples of critically ill patients is increasingly reported even without canonical immunosuppression. The main aim of this study was to better understand the significance of herpesviruses finding in lower respiratory tract of ARDS patients hospitalized in ICU. The presence of this group of herpesviruses, in addition to the research of influenza viruses and other common respiratory viruses, was investigated in respiratory samples from 54 patients hospitalized in ICU, without a known microbiological causative agent. Moreover, the immunophenotype of each patient was analyzed. Herpesviruses DNA presence in the lower respiratory tract seemed not attributable to an impaired immunophenotype, whereas a significant correlation was observed between herpesviruses positivity and influenza virus infection. A higher ICU mortality was significantly related to the presence of herpesvirus infection in the lower respiratory tract as well as to impaired immunophenotype, as patients with poor outcome showed severe lymphopenia, affecting in particular T (CD3+) cells, since the first days of ICU hospitalization. In conclusion, these results indicate that herpesviruses lower respiratory tract infection, which occurs more frequently following influenza virus infection, can be a negative prognostic marker. An independent risk factor for ICU patients with ARDS is an impaired immunophenotype.
Similar content being viewed by others
Introduction
Human herpes simplex virus 1 (HSV1), human cytomegalovirus (hCMV) and Epstein–Barr virus (EBV) are well-known members of the Herpesviridae family, which are highly prevalent and ubiquitous. Primary infection takes place in the majority of cases early in the life and is followed by a lifelong latent infection, from which reactivation may occur with viral shedding at least in the saliva. The outcome of reactivation strongly depends from the host immunological status. In immunodepressed patients, all these three viruses may cause severe diseases, which may be different depending on the virus and on other factors, including host defences. Mostly, hCMV and also HSV1 may cause severe respiratory diseases, whereas the role of EBV in pneumonia is debated [1]. In addition to a direct involvement of these viruses in respiratory diseases, their detection has been associated with other clinical aspects, which may promote viral reactivation or which outcome may be influenced by viral reactivation.
An increasing number of papers report the presence of HSV1, hCMV and EBV in respiratory samples of critically ill patients even without canonical immunosuppression [1–5]. In patients requiring mechanical ventilation, herpesviruses, mainly HSV1 and hCMV, may be frequently detected from either upper or lower respiratory tract samples [6, 7]. It has been suggested that the presence of HSV1 in the respiratory samples of ICU patients correlates with the duration of tracheal intubation [5]. The detection of HSV1 in the lower respiratory tract of ICU patients is reported with a variable frequency, from 5 to 64 % depending on the population and the diagnostic method used [2, 8, 9]. Moreover, it is not always clear whether the demonstration of HSV1 DNA in lower respiratory tract samples of non-immunocompromised ventilated patients is the consequence of a contamination from mouth or throat or is the result of local viral reactivation [4, 6, 7, 10]. Some studies showed that there was a significant association between an HSV1 viral load >100.000 copies/ml of BAL and admission to the ICU (p < 0.0001), mechanical ventilation (p < 0.001) and death (p < 0.01) [5, 11, 12].
Active hCMV infection, either restricted to the lower respiratory tract or involving both the lower respiratory airways and the systemic compartment, has been shown to occur frequently during critical illness in adult hCMV-seropositive patients [13], and has been associated with prolonged ICU hospitalization, extended periods of mechanical ventilation, higher rates of nosocomial infection and overall mortality [11, 14–17].
The role of EBV presence in respiratory tract of ICU patients is not clear. High degree of variability concerning the prevalence of EBV in BAL samples from patients admitted in ICU is reported in the literature [1, 18–21].
ARDS is today a leading cause of hospitalization in ICU. ARDS and pneumonia are closely related to critically ill patients [22]; however, it is not always identified the etiologic agent. In most cases, bacterial infections are the main causative agent of pulmonary infections that evolve into framework of ARDS; more recently, viral infections, mainly related to influenza viruses, represent a new category of emerging cause of ARDS, and also viruses belonging to other families, in association or not to bacterial infections, may be involved. In still other cases, the causative agent remains unrecognized [23]. Furthermore, the critically ill patients develop a state of immunosuppression, which can promote the onset and exacerbation of viral infections [24].
The aim of this study was to better understand the significance of herpesviruses finding in lower respiratory tract of patients hospitalized in ICU and to assess the diagnostic and prognostic value of these findings. Patients’ characteristics, with attention to their immunological setting, were analyzed together with the virological data.
Materials and methods
Institutional Internal Committee approval was waived for this study as it involved retrospective analysis of anonymous, routinely collected, group data. During the period September 2011–May 2014, 164 patients with diagnosis of ARDS were admitted to ICU (Intensive Care Unit of Emergency Department—Careggi Teaching Hospital, Florence—Italy), from different clinical setting. For 54 out of these 164 patients, the causative agent of ARDS was unknown. The following samples were collected for microbiological analysis:
-
Throat swab (TS) and bronchoalveolar lavage (BAL) sent to general laboratory for research of common germs;
-
TS and BAL sent to virology laboratory for the detection of influenza virus and other respiratory viruses like adenovirus (Adv), parainfluenza viruses 1–4 (PIV 1–4), enterovirus/rhinovirus (EV/RhV), respiratory syncytial virus (RSV), human coronaviruses (hCoV) group I and group II, human metapneumovirus (hMPV) and herpetic viruses.
For each patient, the following data were collected:
-
anamnestic data: age, sex, body mass index (BMI), Charlson comorbidity index (CCI) adjusted for age;
-
data and severity scores at ICU admission: SAPS II at admission, SOFA at admission, GCS at admission, provenience, length of stay before ICU admission;
-
data related to respiratory samples: sampling timing, positivity for influenza viruses RNA as well as for other respiratory viruses genome sequences, HSV1/hCMV/EBV DNA; HSV1/hCMV/EBV viral load in BAL;
-
immunophenotyping analysis at ICU admission;
-
data related to ICU stay: treatment with antiviral, steroid; need for extracorporeal membrane oxygenation (ECMO) support and duration of treatment with ECMO;
-
outcome data: SAPS II at discharge, GCS at discharge, ventilation length of stay (LOS), ICU LOS, ICU mortality, post-ICU LOS, post-ICU mortality.
Clinical sampling and analytical phase
In the study period, 108 samples were analyzed with the aim to look for the presence of herpesviruses in 54 patients.
All clinical samples were collected using standard techniques [25]. The throat swab was obtained with a nylon fiber tip (Copan Eswab™ System) inserted and rotated into the throat of patient. The BAL samples were taken with sterile flexible bronchoscope through the oro-tracheal tube or the tracheal cannula; after the assessment of the tracheal–bronchial tree, 30 ml of sterile saline solution was instilled and picked up in a specimen trap (Covidien Argyle™).
The detection and typing of influenza viruses were achieved as already described, using primers and probe sequence as indicated by the US Centers for Disease Control (CDC) [26]. For the detection of other respiratory viruses, duplex real-time PCR, already described, was used [27].
The detection of HSV1 DNA, hCMV DNA and EBV DNA was performed by in-house assays. The in-house assays here described were already used in the laboratory of virology and had shown a performance comparable with commercial assays, at a lower cost. Any way the results here reported were confirmed by comparison with commercial, validated assays (Realtime Q-PCR kit, ELITech Molecular Diagnostics).
Viral DNA extraction and real-time PCRs
Extraction of viral DNAs from clinical samples was carried out using a commercially available kit (HP PCR Template Preparation Kit, Roche Diagnostics, Milan, Italy).
To detect hCMV, HSV1 and EBV DNA in TS and BAL samples, three real-time PCRs were developed, using primers listed in Table 1. The real-time PCRs were performed using 2X HRM PCR master mix (Qiagen, Valencia, CA, USA). The reaction volume for each amplification was 25 μl (12.5 μl of master mix, 1.75 μl of each primer [10 μM], 5 μl of DNA and H2O to reach the final volume). After initial activation step, 40 cycles of amplification [95 °C for 10 s, 55 °C for 30 s, 72 °C for 10 s (acquiring Green)] were performed. For melting analysis, ramp from 78 to 92 °C was used, rising by 0.1 °C each step. The reaction was performed on Rotor Gene 6000 (Qiagen, Valencia, CA, USA).
All herpesvirus-positive BAL samples were quantified by quantitative real-time PCRs.
To perform the calibration curves, serial dilutions of DNA calibrator for each virus were used. These calibrators consisted of DNA sequences obtained by the cloning the product of the PCR of viral DNA of each virus in the pGEM-T Easy Vector System (Promega, Madison, Wisconsin, USA). The plasmid DNA was purified by QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA, USA).
The analytical sensitivity of all PCRs was determined using serial dilutions of cloned calibrators, quantified by NanoDrop 1000 Spectrophotometer (ThermoScientific, Wilmington, DE, USA). The real-time PCR for EBV was able to detect 300 copies number/ml. The sensitivity of real-time PCR for hCMV and HSV1 was 100 copies number/ml.
As the volumes and other characteristics of BAL samples can vary, each BAL sample was quantitatively analyzed also for the β-globin gene, as described below. Then, the results obtained for each sample were normalized according to the ratio [sample target Ct value × sample β-globin Ct value/mean β-globin Ct value] [28].
Human β-globin gene real-time PCR
The detection of β-globin gene was performed using the primers described in the literature [29]. The sequence of primers was PF GH20 5′-caadttcatccacgttcacc-3′ and PR PC04 5′-gaagagccaaggacaggtac-3′. The real-time PCR was performed using 2X HRM PCR master mix (Qiagen, Valencia, CA, USA). The reaction volume was 25 μl (12.5 μl of master mix, 1.75 μl of each primer [10 μM], 5 μl of DNA and H2O to reach the final volume).
After initial activation step, 40 cycles of amplification [95 °C for 10 s, 55 °C for 30 s, 72 °C for 10 s (acquiring Green)] were performed. For melting analysis, ramp from 80 to 90 °C was used, rising by 0.1 °C each step. The reaction was performed on Rotor Gene 6000 (Qiagen, Valencia, CA, USA).
Flow cytometry analysis
Peripheral blood samples (50 µl) were incubated with the appropriate fluorochrome-conjugated mAbs (anti-CD3, CD4, CD8, CD19, CD16, CD56 and HLA-DR) at room temperature for 15 min; red blood cells were then lysed by an appropriate lysing solution (500 µl, BD Biosciences) and acquired with a BDLSR II flow cytometer according to manufacturer’s instructions (BD Biosciences). At least 50.000 cells were acquired and analyzed by using the FACS Diva software (BD Biosciences) [30–32].
Statistical analysis
The descriptive analysis is presented as mean and percentage frequencies. The mean values of the groups were compared using the Student’s t test for numeric values and Chi-square test for ordinary variables. The analysis of variance (ANOVA) was used for comparison of the four groups divided according to positivity for viral infections. We created a logistic model to search for variables predictors of death and a receiver operating characteristic (ROC) curve to identify the cutoff of SAPS II and CD3+ that discriminate for mortality.
A p value <0.05 is considered statistically significant.
For statistical analysis and graphic representation of data were used Software Microsoft Excel 2007©, Graph Pad Prism 6.1© and PASW 17.0© for Windows (IBM Corporation, Armonk, NY, USA).
Results
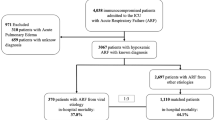
This study includes 54 patients who, since September 2011 to May 2014, were admitted to ICU, from different clinical settings (other ICUs in 68.5 %, ward in 11.1 % and Emergency Department in 20.3 %; mean hospital stay pre-ICU admission was 4.55 ± 6.65 days). This group represents 33.9 % of all patients admitted in ICU with diagnosis of ARDS, without a known microbiological causative agent; within 48 h after ICU admission, clinical samples from these patients were sent to the laboratory for the detection both bacterial and viral infections and for immunophenotyping analysis to assess the immunological status of patients.
The descriptive analysis of the entire sample of patients is illustrated in Table 2.
In 48.1 % of cases, patients required extracorporeal membrane oxygenation (ECMO) for severe ARDS, with hypoxia and/or hypercapnia unresponsive to conventional treatment. The ECMO LOS was on average 15.83 ± 13.13 days.
One hundred and eight clinical samples from upper and lower respiratory tract from the 54 ICU patients were analyzed to detect influenza and other respiratory viruses and a group of herpesviruses (EBV, hCMV and HSV1). These samples were obtained in 96 % of patients the same day of admission or 24–48 h after ICU admission. Nineteen patients were infected by an influenza virus as demonstrated by the detection of viral genome in both upper and lower respiratory samples: 17 were positive for influenza A(H1N1)pdm09 virus, 1 for influenza A(H3N2) virus and 1 for type B virus. Bacterial coinfections were present in 8 patients with laboratory confirmed influenza and in 20 influenza-negative patients (data not shown). Among influenza-negative patients, one adenovirus infection was demonstrated in BAL, one RhV was present in both upper and lower respiratory tract, and one hMpV was demonstrated only in the upper respiratory tract sample.
A total of 35 patients (65 %) were positive for one or more herpesviruses in at least one respiratory sample (18 TF only, 7 BAL only, 10 both samples). Thus, altogether, herpesviruses were present in BAL from 17 patients (31 %) and in TS from 28 patients (52 %). EBV was detected in 23 out of 54 patients (43 %), either as a single infection or as mixed infection. In only 5 patients (9 %), EBV DNA was demonstrated in BAL samples. In 3 cases, it was present as a single infection and in the two other as a mixed infection. hCMV was detected in 15 patients (28 %), either as single (in 2 patients) or mixed infection (in 13 patients). In seven patients (13 %), hCMV DNA was demonstrated in BAL samples. As regards HSV1, viral DNA was detected in 15 patients (28 %). In 9 of these (17 %), it was present in BAL.
In addition, as BAL represents a sample more suggestive of lower respiratory tract infection and/or of more invasive infection/reactivation, to understand better the significance of herpesviruses presence in this site, herpesviruses DNA load in BAL samples was assessed by quantitative real-time PCRs. EBV DNA viral load in BAL samples varied between traces (not quantifiable) in one sample to 1 × 109 copies number/ml in another sample with a median value of 103 copies/ml. Altogether, EBV DNA load (mean ± SD) was 21,254 ± 44,023. The range of hCMV DNA load varied between traces (not quantifiable) in one sample to 1 × 105 copies number/ml with a median value of 103 copies/ml also in this case. Altogether, hCMV DNA load (mean ± SD) was 4240 ± 2619. The load of HSV1 in BAL samples varied between 10 copies number/ml in one sample only and 109 copies number/ml with a median value of 107. Altogether, HSV1 DNA load (mean ± SD) was 142,629,452 ± 422,767,741.
According to herpetic viral infection positivity, patients were divided into 4 groups: group of hCMV-positive patients (n = 7); group of EBV-positive patients (n = 5); group of HSV1-positive patients (n = 9); and group of herpesvirus-negative patients (n = 37).
Patients positive for more than 1 herpesvirus have been included in more than one group.
The analysis of the 4 groups is shown in Table 3.
There were no statistically significant differences in the medical history data, the severity score values at ICU admission and the provenience data. No statistically significant difference in corticosteroid treatment and in the need for extracorporeal treatment was observed. Outcome data showed no statistically significant differences, except for a higher mortality in ICU in patients with herpetic viral infection (hCMV group: 71.4 %, HSV1 group: 55.5 %, EBV group: 40.0 %, herpesvirus-negative group: 21.6 %; p < 0.05).
A significant correlation emerged between influenza virus infection and herpetic viruses coinfection (p < 0.05). All patients with influenza positivity were treated with oseltamivir. In patients with persistent influenza infection, zanamivir was added.
Dividing patients into two groups, based on the positivity for influenza virus, no correlation emerged between influenza infection and ICU mortality. The statistically significant data observed are reported in Table 4: patients with influenza infection showed higher incidence of herpesviruses coinfection in comparison with patients without influenza (52.6 vs 20.0 %; p = 0.01); SAPS II demission score was 11 ± 6.78 for influenza-positive patients, whereas it was 27 ± 11.75 for patients without influenza. In addition, the CD3+ percentage was 68.23 ± 7.24 for influenza-positive patients and 60.37 ± 16.28 for influenza-negative patients. This observation is in agreement with the presence of lymphocytosis as a risk factor for ICU admission in laboratory confirmed influenza patients [33].
An additional analysis was performed by dividing patients into two groups on the basis of ICU mortality: the group of survivors included 37 patients discharged from the ICU; the group of non-survivors included 17 patients died in ICU.
The statistically significant data are shown in Table 5.
ICU mortality was significantly associated with herpesviruses infection in the lower respiratory tract. In fact, 53 % of herpesviruses infected patients died in ICU compared to 22 % in herpesviruses non-infected patients. Only few patients with laboratory confirmed herpesviruses infection were treated with acyclovir/ganciclovir and despite the treatment they died; however, the small number of observations and the lack of virological monitoring does not allow us to tray any conclusion. Several immunological parameters were significantly impaired in the group of patients ICU died. In particular, a clear reduction in circulating lymphocytes was evident in this group when compared to the group of patients discharged from ICU. The cell reduction involves all lymphocytes populations: T (CD3+), B (CD19+) and NK cells (CD3CD16/56+). These data can account for both extravasation of cells that are recruited in inflamed organs and for cell apoptosis that typically affects hyper-activated lymphocytes. To evaluate the influence on ICU mortality of viral coinfections, patients were divided into 4 groups depending on the presence of influenza and/or herpetic infection. ANOVA analysis showed no statistically significant difference in ICU mortality. In the group (n = 10) with presence of both infections (herpes and influenza), ICU mortality was 40 %; in patients with only influenza positivity (n = 9), ICU mortality was 22.3 %, while in patients with only herpetic positivity (n = 7), it was 71.4 %; ICU mortality was 21.5 % in patients with no one viral positivity (n = 28) (p = 0.30). Even if these differences are not significant, these data add further evidence to the association of ICU mortality with herpesviruses infection, whereas ICU mortality in patients with only influenza infection is similar to that of patients negative for both viruses.
To better investigate the variables most associated with mortality, we built a logistic model with SAPS II, herpesviruses positivity and total CD3 value. Candidate variables were chosen as those statistically significant and/or clinically relevant to the outcome.
Table 6 shows that the only variable significantly associated with mortality is herpetic infection; this indicates that herpesviruses positivity is an independent predictor of death. Moreover, we researched a cutoff value for SAPS II and CD3+ (Table 7; Figs. 1, 2).
The logistic regression is slightly over-fitted, but the Hosmer–Lemenshow test is not significant (p = 0.149), suggesting a good calibration of the model.
Discussion and conclusions
ARDS is a relevant disease today, affecting patients of all ages that may require admission to intensive care unit. The mortality for this pathology is still high, despite the implementation of specific therapies in recent years. In patients with ARDS, bacterial infections are prevalent; however, there are no enough studies that highlight the presence of viral etiology. Among respiratory viruses, influenza A viruses, above all of the subtype (H1N1)pdm09, may be associated with ARDS, as it became evident during and after the last influenza pandemic. Some studies report the frequent presence of herpesviruses in respiratory samples of patients with ARDS [1–7, 34]. However, the significance of this positivity is still debated.
This report concerns a group of 54 patients admitted to ICU because of ARDS with unknown causative agent; 19 of them were infected by influenza virus, as demonstrated by the detection of viral RNA in both upper and lower respiratory tract samples. Instead, two other patients, influenza negative, were positive in the BAL for RhV and AdV, respectively. This study confirms that influenza viruses, mainly the H1N1 pandemic subtype, are frequently related to ARDS requiring ICU hospitalization, whereas other common respiratory viruses showed to be involved only sporadically. In 10 of 19 influenza-positive patients, also the DNA of one or more herpesvirus was present in the BAL, whereas no coinfection with herpesviruses was found in the patients with RhV or AdV infection. Thus, in BAL of 7 patients, DNA of herpesviruses alone was found. These data indicate that in 28/54 patients viral infections seem to be involved in ARDS. However, the number of respiratory virus-positive patients could be underestimated because of the time elapsed between the onset of symptoms and the ICU hospitalization. In addition, this study concerned the common respiratory viruses, whereas others like bocavirus and mimivirus were not included. Moreover, it is possible that other already unknown viruses exist. Data on other causative agents as bacteria or fungi have not been considered in this study. In addition to the detection of a direct viral cause of ARDS, this study highlights the existence of some interaction among different viruses and also among viruses, immune status and outcome of ARDS. In fact, a significant correlation was observed between influenza infection and herpesviruses reactivation, demonstrated by the detection of the viral DNA in the BAL. This observation could suggest that the respiratory mucosa damage caused by influenza virus replication can trigger herpesviruses reactivation. As regards each herpesvirus searched in the respiratory tract of the patients included in this study, EBV was the more frequently detected (43 %), whereas both hCMV and HSV1 were present in the respiratory tract of 28 % of patients. However, in BAL EBV DNA was found in 5 patients only (9 %), and hCMV DNA and HSV1 DNA were found in 7 (13 %) and 9 (17 %) patients, respectively. The frequency and DNA load of HSV1 in BAL samples were higher than that of hCMV and EBV, and in 6 patients, it was higher than 100,000 copies/ml, a value that is reported in the literature [5] as related to higher mortality. These results are in agreement with those of Tachikawa [35] who reported that reactivation of HSV1 was predominantly observed in intubated patients regardless of their immune status, whereas reactivation of hCMV and EBV was rare in immunocompetent patients.
Herpesviruses reactivation, as could be inferred by the detection of viral DNA in BAL, was not significantly associated with impaired immunophenotype, whereas it showed to be related to ICU mortality. In particular, the highest ICU mortality was observed among patients with hCMV reactivation, followed by those with HSV1 reactivation and then by those with EBV reaction. As regards the role of each herpesvirus here considered, the small number of data for each virus does not allow to draw a definitive conclusion. Altogether, it seems that EBV may be involved in ARDS like the two other herpesviruses, with a slightly lower frequency. Furthermore, the data analyzed in this study indicate that ICU mortality was significantly related to an impaired immunophenotype as patients with poor outcome showed severe lymphopenia, affecting in particular T (CD3+) cells, since the first days of ICU hospitalization.
In the present study, for the first time, as far as we know, several factors, like respiratory viral infections, respiratory infection/reactivation by some herpesviruses and immune status of the patients, have been considered and analyzed together. The results obtained, even if on a small number of patients, suggest that in a situation such complex as ARDS and in its outcome these factors may act at same time and synergistically: among these, viral respiratory infection, mainly by influenza A(H1N1)pdm09, herpesviruses reactivation (more frequently HSV1, hCMV and also EBV), which may be triggered by the influenza infection, and immune factors (as impaired immunophenotype).
This study has several limitations which are in part related to its observational nature and the scanty samples number. It emphasizes the importance of BAL analysis, whereas the analysis of viremia was performed only in few patients so that we were not able to afford a systematic analysis of these data, which must be implemented in future studies. In addition, it lacks dynamic data on herpesviruses infection, like resolution or persistence of viral infections. In addition, the usefulness of acyclovir/ganciclovir administration needs to be better studied.
The data obtained imply that in ARDS ICU patients—influenza virus laboratory diagnosis should be performed more frequently and as soon as possible; herpesviruses lower respiratory tract infection should monitored, together with the immunological evaluation. This could allow for a timely anti-influenza treatment which could decrease the influenza virus damage on the respiratory mucosa and eventually decrease the probability of herpesviruses reactivation. Data deriving from the study of the immunological setting suggest that the evaluation of the immunophenotype is essential in order to improve the risk stratification in patients affected by systemic virus infection.
References
Friedrichs I, Bingold T, Keppler OT, Pullmann B, Reinheimer C, Berger A (2013) Detection of herpesvirus EBV DNA in the lower respiratory tract of ICU patients: a marker of infection of the lower respiratory tract? Med Microbiol Immunol 202:431–436
Luyt CE, Combes A, Deback C, Aubriot-Lorton MH, Nieszkowska A, Trouillet JL, Capron F, Agut H, Gibert C, Chastre J (2007) Herpes simplex virus lung infection in patients undergoing prolonged mechanical ventilation. Am J Respir Crit Care Med 175:935–942
Smith CA, Conroy LT, Pollock M, Ruddy J, Binning A, Mc Cruden EAB (2010) Detection of herpes viruses in respiratory secretions of patients undergoing artificial ventilation. J Med Virol 82:1406–1409
De Vos N, Van Hoovels L, Vankeerberghen A, Van Vaerenbergh K, Boel A, Demeyer I, Creemers L, De Beenhouwer H (2009) Monitoring of herpes simplex virus in the lower respiratory tract of critically ill patients using real-time PCR: a prospective study. Clin Microbiol Infect 15:358–363
Costa C, Sidoti F, Saldan A, Sinesi F, Balloco C, Simeone S, Lorusso M, Mantovani S, Merlino C, Solidoro P, Cavallo R (2012) Clinical impact of HSV-1 detection in the lower respiratory tract from hospitalized adult patients. Clin Microbiol Infect 18(8):E305–307
Prellner T, Flamholc L, Haidl S, Lindholm K, Widell A (1992) Herpes simplex virus the most frequently isolated pathogen in the lungs of patients with severe respiratory distress. Scand J Infect Dis 24:283–292
Tuxen DV, Cade JF, McDonald MI, Buchanan MRC, Clark RJ, Pain MCF (1982) Herpes simplex virus from the lower respiratory tract in adult respiratory distress syndrome. Am Rev Respir Dis 126:416–419
Bruynseels P, Jorens PG, Demey HE, Goossens H, Pattyn SR, Elseviers MM, Weyler J, Bossaert LL, Mentens Y, Ieven M (2003) Herpes simplex virus in the respiratory tract of critical care patients: a prospective study. Lancet 362:1536–1541
Oud L et al (2006) Comment on: ‘nosocomial viral ventilator-associated pneumonia in the intensive care unit’ by Daubin et al. Intensive Care Med 32:613
Ramsey PG, Fife KH, Hackman RC, Meyers JD, Corey L (1982) Herpes simplex virus pneumonia: clinical, virologic, and pathologic features in 20 patients. Ann Intern Med 97:813–820
Chiche L, Forel JM, Roch A, Guervilly C, Pauly V, Allardet-Servent J, Gainnier M, Zandotti C, Papazian L (2009) Active cytomegalovirus infection is common in mechanically ventilated medical intensive care unit patients. Crit Care Med 37:1850–1857
Frobert E, Billaud G, Casalegno JS, Eibach D, Goncalves D, Robert JM, Lina B, Morfin F (2013) The clinical interest of HSV1 semi-quantification in bronchoalveolar lavage. J Clin Virol 58:265–268
Blanquer J, Chilet M, Benet I, Aguilar G, Mun˜ oz-Cobo B, Tellez A, Costa E, Bravo D, Navarro D (2011) Immunological insights into the pathogenesis of active CMV infection in non-immunosuppressed critically ill patients. J Med Virol 83:1966–1971
Kalil AC, Florescu DF (2009) Prevalence and mortality associated with cytomegalovirus infection in non-immunosuppressed patients in the intensive care unit. Crit Care Med 37:2350–2358
Limaye AP, Boeckh M (2010) CMV in critically ill patients: pathogen or bystander? Rev Med Virol 20:372–379
Ziemann M, Sedemund-Adib B, Reiland P, Schmucker P, Hennig H (2008) Increased mortality in long-term intensive care patients with active cytomegalovirus infection. Crit Care Med 36:3145–3150
Von Muller L, Klemm A, Weiss M, Schneider M, Suger-Wiedeck H, Durmus N, Hampl W, Mertens T (2006) Active cytomegalovirus infection in patients with septic shock. Emerg Infect Dis 12:1517–1522
Young LS, Rickinson AB (2004) Epstein–Barr virus:40 years on. Nat Rev Cancer 4:757–768. doi:10.1038/nrc1452
Gooskens J, Templeton KE, Claas EC, van Bussel MJ, Smit VT, Kroes AC (2007) Quantitative detection of herpes simplex virus DNA in the lower respiratory tract. J Med Virol 79:597–604
Costa C, Elia M, Astegiano S, Sidoti F, Terlizzi ME, Solidoro P, Botto S, Libertucci D, Bergallo M, Cavallo R (2008) Quantitative detection of Epstein–Barr virus in bronchoalveolar lavage from transplant and nontransplant patients. Transplantation 86:1389–1394. doi:10.1097/TP.0b013e3181890415
Costa C, Delsedime L, Solidoro P, Curtoni A, Bergallo M, Libertucci D, Baldi S, Rinaldi M, Cavallo R (2010) Herpesviruses detection by quantitative real-time polymerase chain reaction in bronchoalveolar lavage and transbronchial biopsy in lung transplant: viral infections and histopathological correlation. Transplant Proc 42:1270–1274. doi:10.1016/j.transproceed.2010.03.086
Bauer TT, Ewig S, Rodloff AC, Müller EE (2006) Acute respiratory distress syndrome and pneumonia: a comprehensive review of clinical data. Clin Infect Dis 43(6):748–756
Bauer TT, Valencia M, Badia JR, Ewig S, González J, Ferrer M, Torres A (2005) Respiratory microbiology patterns within the first 24 h of ARDS diagnosis: influence on outcome. Chest 128(1):273–279
Payen D, Faivre V, Lukaszewicz AC, Losser MR (2000) Assessment of immunological status in the critically ill. Minerva Anestesiol 66(5):351–357
Wimberley LL, Falling J, Bartlett JG (1979) A fiberoptic bronchoscopy technique to obtain uncontaminated lower airway secretions for bacterial culture. Am Rev Respir Dis 119:337–343
The WHO Collaborating Centre for influenza at CDC Atlanta, United States of America. CDC protocol of real time RT PCR for Swine Influenza A(H1N1) 2009. http://www.who.int/csr/resources/publications/swineflu/CDCrealtimeRTPCRprotocol20090428.pdf. Accessed 29 April 2009
Arvia R, Corcioli F, Ciccone N, Della Malva N, Azzi A (2015) Detection of 12 respiratory viruses by duplex real time PCR assays in respiratory samples. Mol Cell Probes 29:408–413
Bouscambert Duchamp M, Casalegno JS, Gillet Y, Frobert E, Bernard E, Escuret V, Billaud G, Valette M, Javouhey E, Lina B, Floret D, Morfin F (2010) Pandemic A(H1N1) 2009 influenza virus detection by real time RT-PCR:is viral quantification useful? Clin Microbiol Infect 16(4):317–321. doi:10.1111/j.1469-0691.2010.03169
Bauer HM, Ting Y, Greer CE, Chambers JC, Tashiro CJ, Chimera J, Reingold A, Manos MM (1991) Genital human papillomavirus infection in female university students as determined by a PCR-based method. JAMA 265(4):472–477
Shi SJ, Li H, Liu M, Liu YM, Zhou F, Liu B, Qu JX, Cao B (2015) Mortality prediction to hospitalized patients with influenza pneumonia:PO2/FiO2 combined lymphocyte count is the answer. Clin Respir J. doi:10.1111/crj.12346
Drewry AM, Samra N, Skrupky LP, Fuller BM, Compton SM, Hotchkiss RS (2014) Persistent lymphopenia after diagnosis of sepsis predicts mortality. Shock 42(5):383–391
Boonnak K, Vogel L, Feldmann F, Feldmann H, Legge KL, Subbarao K (2014) Lymphopenia associated with highly virulent H5N1 virus infection due to plasmacytoid dendritic cell-mediated apoptosis of T cells. J Immunol 192(12):5906–5912
Rovina N, Erifaki M, Katsaounou P, Lyxi G, Koutsoukou A, Koulouris GN, Alchanatis M (2014) Subjects Hospitalized With the 2009 Pandemic Influenza A (H1N1) Virus in a Respiratory Infection Unit: Clinical Factors Correlating With ICU Admission. Respir Care 59(10):1560–1568. doi:10.4187/respcare.03049
Lepiller Q, Sueur C, Solis M, Barth H, Glady L, Lefebvre F, Fafi-Kremer S, Schneider F, Stoll-Keller F (2015) Clinical relevance of herpes simplex virus viremia in Intensive Care Unit patients. J Infect 71(1):93–100. doi:10.1016/j.jinf.2015.02.013
Tachikawa R, Tomii K, Seo R, Nagata K, Otsuka K, Nakagawa A, Otsuka K, Hashimoto H, Watanabe K, Shimizu N (2013) Detection of herpes viruses by multiplex and real-time polymerase chain reaction in bronchoalveolar lavage fluid of patients with acute lung injury or acute respiratory distress syndrome. Respiration 87:279–286. doi:10.1159/000355200
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bonizzoli, M., Arvia, R., di Valvasone, S. et al. Human herpesviruses respiratory infections in patients with acute respiratory distress (ARDS). Med Microbiol Immunol 205, 371–379 (2016). https://doi.org/10.1007/s00430-016-0456-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00430-016-0456-z