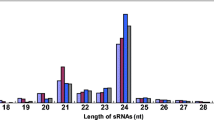
Abstract
Main conclusion
Deep sequencing identified 497 conserved and 559 novel miRNAs in wheat, while degradome analysis revealed 701 targets genes. QRT-PCR demonstrated differential expression of miRNAs during stages of leaf rust progression.
Bread wheat (Triticum aestivum L.) is an important cereal food crop feeding 30 % of the world population. Major threat to wheat production is the rust epidemics. This study was targeted towards identification and functional characterizations of micro(mi)RNAs and their target genes in wheat in response to leaf rust ingression. High-throughput sequencing was used for transcriptome-wide identification of miRNAs and their expression profiling in retort to leaf rust using mock and pathogen-inoculated resistant and susceptible near-isogenic wheat plants. A total of 1056 mature miRNAs were identified, of which 497 miRNAs were conserved and 559 miRNAs were novel. The pathogen-inoculated resistant plants manifested more miRNAs compared with the pathogen infected susceptible plants. The miRNA counts increased in susceptible isoline due to leaf rust, conversely, the counts decreased in the resistant isoline in response to pathogenesis illustrating precise spatial tuning of miRNAs during compatible and incompatible interaction. Stem-loop quantitative real-time PCR was used to profile 10 highly differentially expressed miRNAs obtained from high-throughput sequencing data. The spatio-temporal profiling validated the differential expression of miRNAs between the isolines as well as in retort to pathogen infection. Degradome analysis provided 701 predicted target genes associated with defense response, signal transduction, development, metabolism, and transcriptional regulation. The obtained results indicate that wheat isolines employ diverse arrays of miRNAs that modulate their target genes during compatible and incompatible interaction. Our findings contribute to increase knowledge on roles of microRNA in wheat–leaf rust interactions and could help in rust resistance breeding programs.











Similar content being viewed by others
References
Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18:758–762
Addo-Quaye C, Miller W, Axtell MJ (2009) CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25:130–131
Akdogan G, Tufekci ED, Uranbey S, Unver T (2016) miRNA-based drought regulation in wheat. Funct Integr Genomics 16:221–233
Akpinar BA, Budak H (2016) Dissecting miRNAs in wheat D genome progenitor, Aegilops tauschii. Front Plant Sci 7:606
Akpinar BA, Kantar M, Budak H (2015) Root precursors of microRNAs in wild emmer and modern wheats show major differences in response to drought stress. Funct Integr Genomics 15:587–598
Alptekin B, Budak H (2016) Wheat miRNA ancestors: evident by transcriptome analysis of A, B and D genome donors. Funct Integr Genomics. doi:10.1007/s10142-016-0487-y
Brenchley R, Spannagl M, Pfeifer M, Barker GL, D’Amore R et al (2012) Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491:705–710
Budak H, Akpinar BA (2015) Plant miRNAs: biogenesis, organization and origins. Funct Integr Genomics 15:523–531
Budak H, Khan Z, Kantar M (2014) History and current status of wheat miRNAs using next-generation sequencing and their roles in development and stress. Brief Funct Genomics 14:189–198
Budak H, Kantar M, Balut R, Akpinar BA (2015) Stress responsive miRNAs and isomiRs in cereals. Plant Sci 235:1–13
Chandra S, Singh D, Pathak J, Kumari S, Kumar M, Poddar R, Balyan HS, Gupta PK, Prabhu KV, Mukhopadhyay K (2016) De novo assembled wheat transcriptomes delineate differentially expressed host genes in response to leaf rust infection. PLoS One 11:e0148453
Chapman EJ, Carrington JC (2007) Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet 8:884–896
Chen FX, Zhang N, Zhang S, Wang G, Yin G (2015) Combined small RNA and degradome sequencing reveals novel miRNAs and their targets in the high-yield mutant wheat strain Yunong 3114. PLoS One 10:0137773
Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interaction: shaping the evolution of the plant immune response. Cell 124:803–814
Dean R, Van-Kan JAL, Pretorius ZA, Hammond-Kosack K, Di Pietero A et al (2012) The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13:414–430
Din M, Barozai MYK (2014) Profiling and characterization of eggplant (Solanum melongena L.) microRNAs and their targets. Mol Biol Rep 41:889–894
Dong QH, Han J, Yu HP, Wang C, Zhao MZ, Liu H, Ge AJ, Fang AG (2012) Computational identification of microRNAs in strawberry expressed sequence tags and validation of their precise sequences by miR-RACE. J Heredity 103:268–277
Dvorak J, Dubcovsky J (2007) Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316:1862–1866
Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM et al (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of miRNA genes. PLoS One 2:e219
Fei Q, Zhang Y, Xia R, Meyers BC (2016) Small RNAs add zing to the zig-zag-zig model of plant defenses. Mol Plant Microbe Interact 29:165–169
Feng J, Lin R, Chen J (2013) Alteration of tomato microRNAs expression during fruit development upon Cucumber mosaic virus and Tomato aspermy virus infection. Mol Biol Rep 40:3713–3722
Folkes L, Moxon S, Woolfenden HC, Stocks MB, Szittya G, Dalmay T, Moulton V (2012) PAREsnip: a tool for rapid genome-wide discovery of small RNA/target interactions evidenced through degradome sequencing. Nucleic Acids Res 40:1–10
German MA, Pillay M, Jeong DH, Hetawal A, Luo S et al (2008) Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol 26:941–946
German MA, Luo S, Schroth G, Meyers BC, Green PJ (2009) Construction of parallel analysis of RNA ends. PARE libraries for the study of cleaved miRNA targets and the RNA degradome. Nat Protocol 4:356–362
Gharat SA, Shaw BP (2015) Novel and conserved miRNAs in the halophyte Suaeda maritime identified by deep sequencing and computational predictions using the ESTs of two mangrove plants. BMC Plant Biol 15:301
Gotz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36:3420–3435
Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A (2005) Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res 33:D121–D124
Gupta SK, Charpe A, Koul S, Haque QMR, Prabhu KV (2006) Development and validation of SCAR markers co-segregating with an Agropyron elongatum derived leaf rust resistance gene Lr24 in wheat. Euphytica 150:233–240
Hu G, Rijkenberg FHJ (1998) Scanning electron microscopy of early infection structure formation by Puccinia recondita f. sp. tritici on and in susceptible and resistant wheat lines. Mycol Res 102:391–399
International Wheat Genome Sequencing Consortium (IWGSC) (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 1251788-1-11
Kantar M, Akpınar BA, Valárik M, Lucas SJ, Doležel J, Hernández P, Budak H (2012) Subgenomic analysis of microRNAs in polyploid wheat. Funct Integr Genomics 12:465–479
Karsch-Mizrachi I, Nakamura Y, Cochrane G (2012) The international nucleotide sequence database collaboration. Nucleic Acids Res 40:D33–D37
Katiyar-Agarwal S, Jin H (2010) Role of small RNAs in host-microbe interactions. Annu Rev Phytopathol 48:225–246
Kenan-Eichler M, Leshkowitz D, Tal D, Noor E, Melamed-Bessudo C, Feldman M, Levy AA (2011) Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics 188:263–272
Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42:D68–D73
Kumar D, Kapoor A, Singh D, Satapathy L, Singh AK, Kumar M, Prabhu KV, Mukhopadhyay K (2014a) Functional characterization of a WRKY transcription factor of wheat and its expression analysis during leaf rust pathogenesis. Funct Plant Biol 41:1295–1309
Kumar D, Singh D, Kanodia P, Prabhu KV, Kumar M, Mukhopadhyay K (2014b) Discovery of novel leaf rust responsive microRNAs in wheat and prediction of their target genes. J Nucleic Acids 2014:article ID 570176. doi:10.1155/2014/570176
Kurtoglu KY, Kantar M, Lucas SJ, Budak H (2013) Unique and conserved microRNAs in wheat chromosome 5D revealed by next-generation sequencing. PLoS One 8:e6980
Kurtoglu KY, Kantar M, Lucas SJ, Budak H (2014) New wheat microRNA using whole-genome sequence. Funct Integr Genomics 14:363–379
Li YF, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, Axtell MJ, Zhang W, Sunkar R (2010) Transcriptome-wide identification of microRNA targets in rice. Plant J 62:742–759
Li Y, Lu YG, Shi Y, Wu L, Xu YJ et al (2014) Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol 164:1077–1092
Lipka V, Kwon C, Panstruga R (2007) SNARE-ware: the role of SNARE-domain proteins in plant biology. Annu Rev Cell Develop Biol 23:147–174
Liu H, Qin C, Chen Z, Zuo T, Yang X et al (2014) Identification of miRNAs and their target genes in developing maize ears by combined small RNA and degradome sequencing. BMC Genom 15:25
Luan Y, Wang W, Liu P (2014) Identification and functional analysis of novel and conserved microRNA in tomato. Mol Biol Rep 41:5385–5394
Lucas SJ, Budak H (2012) Sorting the wheat from the chaff: identifying miRNAs in genomic survey sequences of Triticum aestivum chromosome 1AL. PLoS One 7:e40859
McIntosh RA, Pretorius ZA (2011) Borlaug Global Rust Initiative provides momentum for wheat rust research. Euphytica 179:1–2
Meng F, Liu H, Wang K, Liu L, Wang S, Zhao Y, Yin J, Li Y (2013) Development-associated microRNAs in grains of wheat (Triticum aestivum L.). BMC Plant Biol 13:140
Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D et al (2008) Criteria for annotation of plant microRNAs. Plant Cell 20:3186–3190
Miller WA, Shen R, Staplin W, Kanodia P (2016) Noncoding RNAs of plant viruses and viroids: sponges of host translation and RNA interference machinary. Mol Plant Microbe Interact 29(3):156–164
Morin RD, Aksay G, Dolgosheina E, Ebhardt HA, Magrini V, Mardis ER, Sahinalp SC, Unrau PJ (2008) Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res 18:571–584
Moxon S, Schwach F, Dalmay T, Maclean D, Studholme DJ, Moulton V (2008) A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics 24:2252–2253
Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Vionnet O, Johns JDG (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312:436–439
Pandey R, Joshi G, Bhardwaj AR, Agarwal M, Katiyar-Agarwal S (2014) A comprehensive genome-wide study on tissue-specific and abiotic stress-specific miRNAs in Triticum aestivum. PLoS One 9:e95800
Prufer K, Stenzel U, Dannemann M, Green RE, Lachmann M, Kelso J (2008) PatMaN: rapid alignment of short sequences to large databases. Bioinformatics 24:1530–1531
Shah J (2005) Lipids, lipases, and lipid-modifying enzymes in plant disease resistance. Annu Rev Phytopathol 43:229–260
Shu Y, Liu Y, Song L, Zhang J, Guo C (2016) Genome-wide investigation of microRNAs and their targets in response to freezing stress in Medicago sativa L., based on high-throughput sequencing. G3-Genes/Genomics/Genetics 6:755–765
Singh D, Bhaganagare G, Prabhu KV, Gupta PK, Mukhopadhyay K (2012) Targeted spatio-temporal expression based characterization of state of infection and time-point of maximum defense in wheat NILs during leaf-rust infection. Mol Biol Rep 39:9373–9382
Stocks MB, Moxon S, Mapleson D, Woolfenden HC, Mohorianu I, Folkes L, Schwach F, Dalmay T, Moulton V (2012) The UEA sRNA workbench: a suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics 28:2059–2061
Sun F, Guo G, Du J, Guo W, Peng H, Ni Z, Sun Q, Yao Y (2014) Whole genome discovery of miRNAs and their targets in wheat, Triticum aestivum L. BMC Plant Biol 14:142
Sunkar R, Zhou X, Zheng Y, Zhang W, Zhu JK (2008) Identification of novel and candidate miRNAs in rice by high-throughput sequencing. BMC Plant Biol 8:25
Sunkar R, Li YF, Jagadeeswaran G (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci 17:196–203
Tagami Y, Inaba N, Kutsuna N, Kurihara Y, Watanabe Y (2007) Specific enrichment of miRNAs in Arabidopsis thaliana infected with Tobacco mosaic virus. DNA Res 14:227–233
Tanaka T, Kobayashi F, Joshi GP, Onuki R, Sakai H et al (2013) Next-Generation survey sequencing and the molecular organization of wheat chromosome 6B. DNA Res 21:103–114
Tang ZH, Zhang LP, Xu CG, Yuan SH, Zhang FT et al (2012) Uncovering small RNA mediated responses to cold stress in a wheat thermosensitive genic male-sterile line by deep sequencing. Plant Physiol 159:721–738
Weiberg A, Wang M, Lin FM, Zhao H, Zhang Z, Kaloshian I, Huang HD, Jin H (2013) Fungal small RNAs suppress RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342:118–123
Weiberg A, Wang M, Bellinger M, Jin H (2014) Small RNAs: a new paradigm in plant–microbe interactions. Annu Rev Phytopathol 52:495–516
Xin M, Wang Y, Yao Y, Xic C, Peng H et al (2010) Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol 10:123
Xu T, Wang Y, Liu X, Lv S, Feng C, Qi M, Li T (2015) Small RNA and degradome sequencing reveals microRNAs and their targets involved in tomato pedicel abscission. Planta 242:963–984
Ye J, Fang L, Zheng H, Zhang Y, Chen J et al (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34:W293–W297
Yu Y, Wu G, Yuan H, Cheng L, Zhao D et al (2016) Identification and characterization of miRNAs and targets in flax (Linum usitatissimum) under saline, alkaline, and saline-alkaline stresses. BMC Plant Biol 16:24
Zhang B, Pan XP, Cobb GP, Anderson TA (2006) Plant microRNA: a small regulatory molecule with big impact. Dev Biol 289:3–16
Zhao YT, Wang M, Wang ZM, Fang RX, Wang XJ, Jia YT (2015) Dynamic and coordinated expression changes of rice small RNAs in response to Xanthomonas oryzae pv. oryzae. J Genet Genomics 42:625–637
Acknowledgments
We are thankful to Centre of Excellence, Technical Education Quality Improvement Program-II, (Grant No. NPIU/TEQIP II/FIN/31/158) for providing financial support and BTISNet SubDIC (BT/BI/04/065/04) for providing facilities for bioinformatics analyses. D. K. is grateful to the Council of Scientific and Industrial Research [9/554 (0026) 2010-EMR-I] and SD to DST INSPIRE (IF140725) for fellowships. We thank Mr. Saket Chandra of BIT, Mesra, and Dr. Fritz Thümmler, CEO, vertis Biotechnolgie AG, Germany, for helpful discussions and Dr Sunil K Mukherjee of ICGEB New Delhi for critical suggestions on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1 Analysis of sRNA samples on a Shimadzu MultiNA Microchip Electrophoresis System. a small RNA samples, M = RNA ladder. b PCR-amplified cDNA, M = 100 bp ladder. c Fractionated and amplified cDNA pool prepared for Illumina sequencing, M = 100 bp ladder. The miRNA library names are mentioned on top of respective lanes
Figure S2 Analysis of RNA samples for degradome preparation on a Shimadzu MultiNA Microchip Electrophoresis System. a total RNA samples, M = RNA ladder. b PCR-amplified cDNA, M = 100 bp ladder. The degradome library names are mentioned on top of respective lanes
Figure S3 Polyacrylamide (6 %) Gel analysis of the MmeI-digested cDNA. The 42-bp-long-released 5′ cDNA fragments are marked. M = 20 bp ladder. The library names are mentioned on top of the respective lanes
Figure S4 Analysis of the PCR-amplified 131-bp-long MmeI fragments. M = 100 bp ladder. The library names are mentioned on top of the respective lanes
Figure S5 Target ‘t’-plots. a Degradome D1. b Degradome D2. c Degradome D3. d Degradome D1
Figure S6 GO terms of the target genes identified in D1 and their enrichment analysis. (a) Analysis of the targets within the molecular function category. (b) Analysis of the targets within the biological process category. (c) Analysis of the targets within the cellular component category. This analysis was performed using the online tool Blast2GO.
Figure S7 GO terms of the target genes identified in D2 and their enrichment analysis. (a) Analysis of the targets within the molecular function category. (b) Analysis of the targets within the biological process category. (c) Analysis of the targets within the cellular component category. This analysis was performed using the online tool Blast2GO.
Figure S8 GO terms of the target genes identified in D3 and their enrichment analysis. (a) Analysis of the targets within the molecular function category. (b) Analysis of the targets within the biological process category. (c) Analysis of the targets within the cellular component category. This analysis was performed using the online tool Blast2GO.
Figure S9 GO terms of the target genes identified in D4 and their enrichment analysis. (a) Analysis of the targets within the molecular function category. (b) Analysis of the targets within the biological process category. (c) Analysis of the targets within the cellular component category. This analysis was performed using the online tool Blast2GO.
Figure S10 KEGG pathway map of oxidative phosphorylation showing the identified conserved miRNA bdi-miR168 targeting the enzyme NADH dehydrogenase (ID 1.6.5.3, shown in Red arrow)
Table S1 Descriptions of cDNA samples for preparation of small RNA libraries
Table S2 Descriptions of cDNA samples for preparation of degradome libraries
Table S3 List of primers used for Real-Time PCR-based expression profiling of selected miRNAs
Table S4 List of conserved miRNAs and their expression profiles
Table S5a List of novel miRNAs and their expression profiles
Table S5b Novel miRNAs identified using wheat draft chromosome
Table S5c Novel miRNAs identified using ESTs as reference
Table S5d List of differentially expressed miRNAs between S-Mmi and S-PImi
Table S5e List of differentially expressed miRNAs between R-Mmi and R-PImi
Table S6a Identified target genes and their characteristics in S-Mmi libraries vs. D1 libraries
Table S6b Identified target genes and their characteristics in S-PImi libraries vs. D2 libraries
Table S6c Identified target genes and their characteristics in R-Mmi libraries vs. D3 libraries
Table S6d Identified target genes and their characteristics in R-PImi libraries vs. D4 libraries
Table S7 List of pathways identified through the KEGG analysis
Rights and permissions
About this article
Cite this article
Kumar, D., Dutta, S., Singh, D. et al. Uncovering leaf rust responsive miRNAs in wheat (Triticum aestivum L.) using high-throughput sequencing and prediction of their targets through degradome analysis. Planta 245, 161–182 (2017). https://doi.org/10.1007/s00425-016-2600-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-016-2600-9