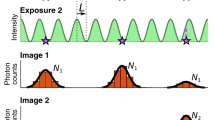
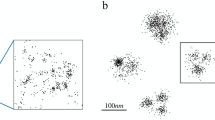
Abstract
Although there are many reconstruction algorithms for localization microscopy, their use is hampered by the difficulty to adjust a possibly large number of parameters correctly. We propose SimpleSTORM, an algorithm that determines appropriate parameter settings directly from the data in an initial self-calibration phase. The algorithm is based on a carefully designed yet simple model of the image acquisition process which allows us to standardize each image such that the background has zero mean and unit variance. This standardization makes it possible to detect spots by a true statistical test (instead of hand-tuned thresholds) and to de-noise the images with an efficient matched filter. By reducing the strength of the matched filter, SimpleSTORM also performs reasonably on data with high-spot density, trading off localization accuracy for improved detection performance. Extensive validation experiments on the ISBI Localization Challenge Dataset, as well as real image reconstructions, demonstrate the good performance of our algorithm.













Similar content being viewed by others
Notes
This is possible because the discretization noise is typically much smaller than the noise from other sources.
References
Abraham AV, Ram S, Chao J, Ward ES, Ober RJ (2009) Quantitative study of single molecule location estimation techniques. Opt Express 17(26):23,352
Andersson SB (2008) Localization of a fluorescent source without numerical fitting. Opt Express 16(23):18,714–18,724
Anscombe FJ (1948) The transformation of Poisson, binomial and negative-binomial data. Biometrika 35(3/4):246–254
Baddeley D (2012) PYME—the python localization microscopy environment. http://code.google.com/p/python-microscopy/. Accessed: 12 Feb. 2014
Bates M, Huang B, Zhuang X (2008) Super-resolution microscopy by nanoscale localization of photo-switchable fluorescent probes. Curr Opin Chem Biol 12(5):505–514
Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313(5793):1642–1645
Boulanger J, Kervrann C, Bouthemy P, Elbau P, Sibarita JB, Salamero J (2010) Patch-based nonlocal functional for denoising fluorescence microscopy image sequences. IEEE Trans Med Imaging 29(2):442–454
Brede N, Lakadamyali M (2012) GraspJ: an open source, real-time analysis package for super-resolution imaging. Optl Nanoscopy 1(1):1–7
Egner A, Geisler C, von Middendorff C, Bock H, Wenzel D, Medda R, Andresen M, Stiel AC, Jakobs S, Eggeling C et al (2007) Fluorescence nanoscopy in whole cells by asynchronous localization of photoswitching emitters. Biophys J 93(9):3285–3290
Fischler MA, Bolles RC (1981) Random sample consensus: a paradigm for model fitting with applications to image analysis and automated cartography. Commun ACM 24(6):381–395
Grüll F, Kirchgessner M, Kaufmann R, Hausmann M, Kebschull U (2011) Accelerating image analysis for localization microscopy with FPGAs. In: Field Programmable Logic and Applications (FPL), 2011 International Conference, pp 1–5
Hedde PN, Fuchs J, Oswald F, Wiedenmann J, Nienhaus GU (2009) Online image analysis software for photoactivation localization microscopy. Nat Methods 6:689–690
Heilemann M (2010) Fluorescence microscopy beyond the diffraction limit. J Biotechnol 149(4):243–251
Heilemann M, van de Linde S, Schüttpelz M, Kasper R, Seefeldt B, Mukherjee A, Tinnefeld P, Sauer M (2008) Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angewandte Chemie Int Ed 47(33):6172–6176
Heilemann M, van de Linde S, Mukherjee A, Sauer M (2009) Super-resolution imaging with small organic fluorophores. Angewandte Chemie Int Ed 48(37):6903–6908
Henriques R, Mhlanga MM (2009) PALM and STORM: what hides beyond the Rayleigh limit? Biotechnol J 4(6):846–857
Henriques R, Lelek M, Fornasiero EF, Valtorta F, Zimmer C, Mhlanga MM (2010) QuickPALM: 3D real-time photoactivation nanoscopy image processing in ImageJ. Nat Methods 7(5):339–340
Herbert A (2012) PeakFit ImageJ plugins for single-molecule light microscopy. http://www.sussex.ac.uk/gdsc/intranet/microscopy/imagej/smlm_plugins. Accessed: 12 Feb. 2014
Högbom JA (1974) Aperture synthesis with a non-regular distribution of interferometer baselines. Astron Astrophys Suppl 15:417
Holden S, Uphoff S, Kapanidis A (2011) DAOSTORM: an algorithm for high-density super-resolution microscopy. Nat Methods 8(4):279
Huang B (2010) Super-resolution optical microscopy: multiple choices. Curr Opin Chem Biol 14(1):10–14
Huang F, Schwartz SL, Byars JM, Lidke KA (2011) Simultaneous multiple-emitter fitting for single molecule super-resolution imaging. Biomed Opt Express 2(5):1377–1393
Izeddin I, Boulanger J, Racine V, Specht C, Kechkar A, Nair D, Triller A, Choquet D, Dahan M, Sibarita J (2012) Wavelet analysis for single molecule localization microscopy. Opt Express 20(3):2081–2095
Kim K, Min J, Carlini L, Unser M, Manley S, Jeon D, Ye J (2013) Fast maximum likelihood high-density low-SNR super-resolution localization microscopy. In: Proceedings 10th International Workshop Sampling Theory and Applications (SampTA’13), pp 285–288
Křížek P, Raška I, Hagen GM (2011) Minimizing detection errors in single molecule localization microscopy. Opt Express 19(4):3226–3235
Lidke K, Rieger B, Lidke D, Jovin T (2005) The role of photon statistics in fluorescence anisotropy imaging. IEEE Trans Image Process 14(9):1237–1245
Min J, Vonesch C, Olivier N, Kirshner H, Manley S, Ye J, Unser M (2013) Continuous localization using sparsity constraints for high-density super-resolution microscopy. In: Proceedings ISBI’13, pp 181–184
Muranyi W, Malkusch S, Müller B, Heilemann M, Kräusslich HG (2013) Super-resolution microscopy reveals specific recruitment of hiv-1 envelope proteins to viral assembly sites dependent on the envelope c-terminal tail. PLoS Pathog 9(2):9(2):e198–e1003
Smith CS, Joseph N, Rieger B, Lidke KA (2010) Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nat Methods 7(5):373–375
Stallinga S, Rieger B (2012) Position and orientation estimation of fixed dipole emitters using an effective Hermite point spread function model. Opt Express 20(6):5896–5921
Stetson PB (1987), DAOPHOT: a computer program for crowded-field stellar photometry. Publications of the Astronomical Society of the Pacific, pp 191–222
Stuurman N (2012) Localization Microscopy MicroManager Plugin. http://micro-manager.org/wiki/Localization_Microscopy. Accessed: 12 Feb. 2014
Thompson RE, Larson DR, Webb WW (2002) Precise nanometer localization analysis for individual fluorescent probes. Biophys J 82(5):2775–2783
Turin G (1960) An introduction to matched filters. IRE Trans Inf Theory 6(3):311–329
Wolter S, Schüttpelz M, Tscherepanow M, van de Linde S, Heilemann M, Sauer M (2010) Real-time computation of subdiffraction-resolution fluorescence images. J Microsc 237(1):12–22
Yu J (2011) Octane ImageJ plugin for super-resolution imaging and single molecule tracking. https://github.com/jiyuuchc/Octane. Accessed: 12 Feb. 2014
Zhu L, Zhang W, Elnatan D, Huang B (2012) Faster STORM using compressed sensing. Nat Methods 9(7):721–723
Acknowledgments
This research was supported by contract research “Methoden für die Lebenswissenschaften” of the Baden-Württemberg Stiftung. We are grateful to Mike Heilemann, Varun Venkataramani and Benjamin Flottmann for providing the raw data of the images presented in this paper, as well as for giving many helpful comments on the algorithm. We also thank the organizers of the ISBI Localization Microscopy Challenge for the permission to use their artificial data.
Author information
Authors and Affiliations
Corresponding author
Additional information
Ullrich Köthe and Frank Herrmannsdörfer have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Köthe, U., Herrmannsdörfer, F., Kats, I. et al. SimpleSTORM: a fast, self-calibrating reconstruction algorithm for localization microscopy. Histochem Cell Biol 141, 613–627 (2014). https://doi.org/10.1007/s00418-014-1211-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-014-1211-4