Abstract
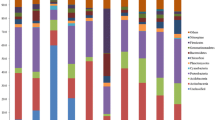
Schirmacher Oasis, Antarctica, is a region with relatively large exposed area and consisted of many freshwater lakes. Nevertheless, only a few studies were done on the bacterial diversity of this region. Hence, this project was undertaken to determine the bacterial community in soil samples collected from the Schirmacher Oasis using the denaturing gradient gel electrophoresis (DGGE) of amplified 16S rDNA fragments. A total of 79 partial 16S rDNA sequences were obtained from the excised DGGE bands, which corresponded to 63 different operational taxonomic units (OTUs) representing bacteria from seven different phyla. The most dominant phyla in descending order were Acidobacteria, Proteobacteria, Bacteroidetes, and Actinobacteria, Planctomycetes, Cyanobacteria and BRC1. There were 5.4 % of unclassified bacteria which cannot be grouped into any of the existing phyla. Eighty-seven percent of the OTUs had highest similarity with the uncultured bacteria from the NCBI GenBank database. Thirty-two percent of the OTUs were similar to bacteria reported in other parts of the Antarctica, while the others were related to bacteria found elsewhere outside the Antarctic.



Similar content being viewed by others
References
Aislabie JM, Chhour KL, Saul DJ, Miyauchi S, Ayton J, Paetzold RF, Balks MR (2006) Dominant bacteria in soils of Marble point and Wright valley, Victoria land, Antarctica. Soil Biol Biochem 38:3041–3056
Aislabie JM, Jordan S, Barker GM (2008) Relation between soil classification and bacterial diversity in soils of the Ross Sea region, Antarctica. Geoderma 144:9–20
Aislabie JAJ, Jordan SJS, Ayton JAJ, Klassen JKJ, Barker GBG, Turner STS (2009) Bacterial diversity associated with ornithogenic soil of the Ross Sea region, Antarctica. Can J Microbiol 55:21–36
Babalola OO, Kirby BM, Roes-Hill L, Cook AE, Cary SC, Burton SG, Cowan DA (2009) Phylogenetic analysis of actinobacterial populations associated with Antarctic Dry Valley mineral soils. Environ Microbiol 11:566–576
Boon N, Windt W, Verstraete W, Top EM (2002) Evaluation of nested PCR–DGGE (denaturing gradient gel electrophoresis) with group-specific 16S rRNA primers for the analysis of bacterial communities from different wastewater treatment plants. FEMS Microbiol Ecol 39:101–112
Borg SG, DePaolo DJ (1994) Laurentia, Australia, and Antarctica as a Late Proterozoic supercontinent: constraints from isotopic mapping. Geology 22:307–310
Bromwich DH, Nicolas JP, Monaghan AJ, Lazzara MA, Keller LM, Weidner GA, Wilson AB (2013) Central West Antarctica among the most rapidly warming regions on earth. Nat Geosci 6:139–145
Chong CW, Annie Tan GY, Wong RCS, Riddle MJ, Tan IKP (2009) DGGE fingerprinting of bacteria in soils from eight ecologically different sites around Casey Station, Antarctica. Polar Biol 32:853–860
Chong CW, Pearce DA, Convey P, Tan GY, Wong R, Tan IKP (2010) High levels of spatial heterogeneity in the biodiversity of soil prokaryotes on Signy Island, Antarctica. Soil Biol Biochem 42:601–610
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009) The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:141–145
Convey P (2010) Terrestrial biodiversity in Antarctica—recent advances and future challenges. Polar Sci 4:135–147
Costello EK, Halloy SR, Reed SC, Sowell P, Schmidt SK (2009) Fumarole-supported islands of biodiversity within a hyperarid, high-elevation landscape on Socompa Volcano, Puna de Atacama, Andes. Appl Environ Microbiol 75:735–747
Cowan DA, Chown SL, Convey P, Tuffin M, Hughes K, Pointing S, Vincent WF (2011) Non-indigenous microorganisms in the Antarctic: assessing the risks. Trends Microbiol 19:540–548
Dalziel IWD (1991) Pacific margins of Laurentia and East Antarctica as a conjugate rift pair; evidence and implications for an Eocambrian supercontinent. Geology 19:598–601
Dunbar J, Ticknor LO, Kuske CR (2000) Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl Environ Microbiol 66:2943–2950
Foong CP, Wong CMVL, Gonzales M (2010) Metagenomic analyses of the dominant bacterial community in the Fildes Peninsula, King George Island (South Shetland Islands). Polar Sci 4:263–273
Fromin N, Hamelin J, Tarnawski S, Roesti D, Jourdain-Miserez K, Forestier N, Teyssier-Cuvelle S, Gillet F, Aragno M, Rossi P (2002) Statistical analysis of denaturing gel electrophoresis (DGE) fingerprinting patterns. Environ Microbiol 4:634–643
Gafan GP, Lucas VS, Roberts GJ, Petrie A, Wilson M, Spratt DA (2005) Statistical analyses of complex denaturing gradient gel electrophoresis profiles. J Clin Microbiol 43:3971–3978
Ganzert L, Lipski A, Hubberten HW, Wagner D (2011) The impact of different soil parameters on the community structure of dominant bacteria from nine different soils located on Livingston Island, South Shetland Archipelago, Antarctica. FEMS Microbiol Ecol 76:476–491
Gregory JW (1901) The work of the national Antarctic expedition. Nature 63:609–612
Head IM, Saunders JR, Pickup RW (1998) Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb Ecol 35:1–21
Holben WE, Jansson JK, Chelm BK, Tiedje JM (1988) DNA probe method for the detection of specific microorganisms in the soil bacterial community. Appl Environ Microbiol 54:703–711
Hughes KA, Convey P, Maslen NR, Smith RIL (2010) Accidental transfer of non-native soil organisms into Antarctica on construction vehicles. Biol Invasions 12:875–891
Kenarova A, Encheva M, Chipeva V, Chipev N, Hristova P, Moncheva P (2013) Physiological diversity of bacterial communities from different soil locations on Livingston Island, South Shetland archipelago, Antarctica. Polar Biol 36:223–233
Li S, Xiao X, Yin X, Wang F (2006) Bacterial community along a historic lake sediment core of Ardley Island, West Antarctica. Extremophiles 10:461–467
Li Z, He L, Miao X (2007) Cultivable bacterial community from South China Sea sponge as revealed by DGGE fingerprinting and 16S rDNA phylogenetic analysis. Curr Microbiol 55:465–472
Mojib N, Bej AK, Hoover R (2008) Diversity and cold adaptation of microorganisms isolated from the Schirmacher Oasis, Antarctica. Proc SPIE 9:70970K–70992K
Muyzer G, Smalla K (1998) Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Van Leeuwenhoek 73:127–141
Muyzer G, Teske A, Wirsen CO, Jannasch HW (1995) Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol 164:165–172
Nakatsu CH (2007) Soil microbial community analysis using denaturing gradient gel electrophoresis. Soil Sci Soc Am J 71:562–571
Nercessian O, Noyes E, Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L (2005) Bacterial populations active in metabolism of C1 compounds in the sediments of Lake Washington, a fresh water lake. Appl Environ Microbiol 71:6885–6899
Newsham KK, Pearce DA, Bridge PD (2010) Minimal influence of water and nutrient content on the bacterial community composition of a maritime Antarctic soil. Microbiol Res 165:523–530
Nichols D, Bowman J, Sanderson K, Nichols CM, Lewis T, McMeekin T, Nichols PD (1999) Development with Antarctic microorganisms: culture collections, bioactivity screening, taxonomy, PUFA production and cold-adapted enzymes. Curr Opin Biotechnol 10:240–246
Peeters K, Verleyen E, Hodgson DA, Convey P, Ertz D, Vyverman W, Willems A (2012) Heterotrophic bacterial diversity in aquatic microbial mat communities from Antarctica. Polar Biol 35:543–554
Polymenakou PN, Lampadariou N, Mandalakis M, Tselepides A (2009) Phylogenetic diversity of sediment bacteria from the southern Cretan margin, Eastern Mediterranean Sea. Syst Appl Microbiol 32:17–26
Porteous LA, Armstrong JL (1991) Recovery of bulk DNA from soil by a rapid, small-scale extraction method. Curr Microbiol 22:345–348
Prabhu Matondkar SG (1986) Microbiological studies in Schirmacher Oasis, Antarctica effect of temperature on bacterial populations. Third Indian Expedition to Antarctica Scientific Report Department of Ocean Development Technical Publication 3:133–147
Rawat SR, Männistö MK, Bromberg Y, Häggblom MM (2012) Comparative genomic and physiological analysis provides insights into the role of Acidobacteria in organic carbon utilization in Arctic tundra soils. FEMS Microbiol Ecol 82:341–355
Rinnan R, Rousk J, Yergeau E, Kowalchuk GA, Bååth E (2009) Temperature adaptation of soil bacterial communities along an Antarctic climate gradient: predicting responses to climate warming. Glob Change Biol 15:2615–2625
Roesch LF, Fulthorpe RR, Pereira AB, Pereira CK, Lemos LN, Barbosa AD, da Costa EM (2012) Soil bacterial community abundance and diversity in ice-free areas of Keller Peninsula, Antarctica. Appl Soil Ecol 61:7–15
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Salwoom L, Tan GY, Tan IKP (2012) Bacterial diversity from Schirmacher Oasis, Antarctica. In: Twenty fourth Indian Antarctic expedition 2004–2005. Ministry of Earth Sciences, Technical Publication No. 22, pp 285–292
Saul DJ, Aislabie JM, Brown CE, Harris L, Foght JM (2005) Hydrocarbon contamination changes the bacterial diversity of soil from around Scott Base, Antarctica. FEMS Microbiol Ecol 53:141–155
Selbmann L, Zucconi L, Ruisi S, Grube M, Cardinale M, Onofri S (2010) Culturable bacteria associated with Antarctic lichens: affiliation and psychrotolerance. Polar Biol 33:71–83
Shivaji S, Rao NS, Saisree L, Reddy GSN, Kumar GS, Bhargava PM (1989) Isolation of Arthrobacter from the soils of Schirmacher Oasis, Antarctica. Polar Biol 10:225–229
Shivaji S, Reddy GSN, Aduri RP, Kutty R, Ravenschlag K (2004) Bacterial diversity of a soil sample from Schirmacher Oasis, Antarctica. Cell Mol Biol 50:525–536
Shivaji S, Kumari K, Kishore KH, Pindi PK, Rao PS, Srinivas TNR, Asthana R, Ravindra R (2011) Vertical distribution of bacteria in a lake sediment from Antarctica by culture-independent and culture-dependent approaches. Res Microbiol 162:191–203
Shravage BV, Dayananda KM, Patole MS, Shouche YS (2007) Molecular microbial diversity of a soil sample and detection of ammonia oxidizers from Cape Evans, McMurdo Dry Valley, Antarctica. Microbiol Res 162:15–25
Smith MC, Bowman JP, Scott FJ, Line MA (2000) Sublithic bacteria associated with Antarctic quartz stones. Antarct Sci 12:177–184
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetic analysis (MEGA) software version 4.0. Mol Biol Evol 24:1569–1599
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Taton A, Grubisic S, Brambilla E, De Wit R, Wilmotte A (2003) Cyanobacterial diversity in natural and artificial microbial mats of Lake Fryxell (McMurdo Dry Valleys, Antarctica): a morphological and molecular approach. Appl Environ Microbiol 69:5157–5169
Teixeira LC, Peixoto RS, Cury JC, Sul WJ, Pellizari VH, Tiedje J, Rosado AS (2010) Bacterial diversity in rhizosphere soil from Antarctic vascular plants of Admiralty Bay, maritime Antarctica. ISME J 4:989–1001
Tsai YL, Olson BH (1992) Detection of low numbers of bacterial cells in soils and sediments by polymerase chain reaction. Appl Microbiol Biotechnol 58:754–757
Vickers CJ (2012) Investigating the physiological and metabolic requirements of the tramway ridge microbial community, Mt Erebus, Antarctica. Doctoral dissertation, University of Waikato, pp 15–85
Vincent WF (2000) Evolutionary origins of Antarctic microbiota: invasion, selection and endemism. Antarct Sci 12:374–385
Vyverman W, Verleyen E, Wilmotte A, Hodgson DA, Willems A, Peeters K, de Vijver BV, Wever AD, Leliaert F, Sabbe K (2010) Evidence for widespread endemism among Antarctic micro-organisms. Polar Sci 4:103–113
Xiao X, Li M, You Z, Wang F (2007) Bacterial communities inside and in the vicinity of the Chinese Great Wall station, King George Island, Antarctica. Antarct Sci 19:11–16
Xue D, Yao HY, Ge DY, Huang CY (2008) Soil microbial community structure in diverse land use systems: a comparative study using biolog, DGGE, and PLFA analyses. Pedosphere 18:653–663
Yergeau E, Bokhorst S, Kang S, Zhou J, Greer CW, Aerts R, Kowalchuk GA (2012) Shifts in soil microorganisms in response to warming are consistent across a range of Antarctic environments. ISME J 6:692–702
Zhou J, Bruns MA, Tiedje JM (1996) DNA recovery from soils of diverse composition. Appl Environ Microbiol 62:316–322
Acknowledgments
We would like to thank the Academic Sciences of Malaysia (ASM) and the Ministry of Science, Technology and Innovation (MOSTI), Malaysia for funding this research project (Research Grant No. 95500-66) and the National Center for Antarctic and Ocean Research (NCAOR), Goa, India, for providing the logistic supports for the sample collections. We would also like to thank Jacqueline Yong for reading and correcting the final draft of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Teo, J.K.C., Wong, C.M.V.L. Analyses of soil bacterial diversity of the Schirmacher Oasis, Antarctica. Polar Biol 37, 631–640 (2014). https://doi.org/10.1007/s00300-014-1463-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00300-014-1463-4