Abstract
Key message
TAS atasiRNA-producing region swapping used one-step, high efficiency, and high fidelity directional TC-cloning. Uniform silencing was achieved without lethality using miRNA trigger- TAS overexpression fusion cassettes to generate 21-nt atasiRNA.
Abstract
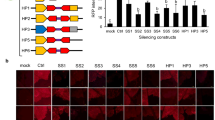
Plant transgenic technologies are very important for basic plant research and biotechnology. Artificial trans-acting small interfering RNA (atasiRNA) represents an attractive platform with certain advantages over other silencing approaches, such as hairpin RNA, artificial microRNA (amiRNA), and virus-induced gene silencing (VIGS). In this study, we developed two types of constructs for atasiRNA-mediated gene silencing in plants. To functionally validate our constructs, we chose TAS1a as a test model. Type 1 constructs had miR173-precursor sequence fused with TAS1a locus driven by single promoter–terminator cassette, which simplified the expression cassette and resulted in uniform gene silencing. Type 2 constructs contained two separate cassettes for miR173 and TAS1a co-expression. The constructs in each type were further improved by deploying the XcmI-based TC-cloning system for highly efficient directional cloning of short DNA fragments encoding atasiRNAs into TAS1a locus. The effectiveness of the constructs was demonstrated by cloning an atasiRNA DNA into the TC site of engineered TAS1a and silencing of CHLORINA 42 (CH42) gene in Arabidopsis. Our results show that the directional TC-cloning of the atasiRNA DNA into the engineered TAS1a is highly efficient and the miR173–TAS1a fusion system provides an attractive alternative to achieve moderate but more uniform gene silencing without lethality, as compared to conventional two separate cassettes for miR173 and TAS locus co-expression system. The design principles described here should be applicable to other TAS loci such as TAS1b, TAS1c, TAS2, or TAS3, and cloning of amiRNA into amiRNA stem-loop.






Similar content being viewed by others
Abbreviations
- amiRNA:
-
Artificial microRNA
- atasiRNA:
-
Artificial trans-acting small interfering RNA
- DCL:
-
Dicer-like protein
- dsRNA:
-
Double-stranded RNA
- sRNA:
-
Small RNA
- siRNA:
-
Small interfering RNA
- TAS :
-
Trans-acting siRNA locus
References
Allen E, Howell MD (2010) miRNAs in the biogenesis of trans-acting siRNAs in higher plants. Semin Cell Dev Biol 21:798–804
Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121:207–221
Axtell MJ (2013) Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64:137–159
Baulcombe DC (2007) Small RNA molecules that silence gene expression are amplified by different mechanisms in nematodes and plants. Science 315:199–200
Baykal U, Zhang Z (2010) Chapter XI: Small RNA-mediated gene silencing for plant biotechnology. In: Catalano AJ (ed) Gene silencing: theory, techniques and applications. Nova Science, New York, pp 255–269
Brodersen P, Voinnet O (2006) The diversity of RNA silencing pathways in plants. Trends Genet 22:268–280
Carbonell A, Takeda A, Fahlgren N, Johnson SC, Cupenrus JT, Carrington JC (2014) New generation of artificial microRNA and synthetic trans-acting small interfering RNA vectors for efficient gene silencing in Arabidopsis. Plant Physiol 165:15–29
Chapman EJ, Carrington JC (2007) Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet 8:884–896
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
de la Luz Gutiérrez-Nava M, Aukerman MJ, Sakai H, Tingey SV, Williams RW (2008) Artificial trans-acting siRNAs confer consistent and effective gene silencing. Plant Physiol 147:543–551
Felippes FF, Weigel D (2009) Triggering the formation of tasiRNAs in Arabidopsis thaliana: the role of microRNA miR173. EMBO Rep 10:264–270
Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J et al (2004) Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res 14:2176–2189
Kovalic D, Kwak JH, Weisblum B (1991) General method for direct cloning of DNA fragments generated by the polymerase chain reaction. Nucleic Acids Res 19:4560
Li JF, Chung HS, Niu Y, Bush J, McCormack M, Sheen J (2013) Comprehensive protein-based artificial microRNA screens for effective gene silencing in plants. Plant Cell 25:1507–1522
Mead DA, Pey NK, Herrnstadt C, Marcil RA, Smith LM (1991) A universal method for the direct cloning of PCR amplified nucleic acid. Biotechnology 9:657–663
Montgomery TA, Yoo SJ, Fahlgren N, Gilbert SD, Howell MD, Sullivan CM et al (2008) AGO1–miR173 complex initiates phased siRNA formation in plants. Proc Natl Acad Sci USA 105:20055–20062
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Park SY, Yin X, Duan K, Gelvin SB, Zhang Z (2014) Heat shock protein 90.1 plays a role in Agrobacterium-mediated plant transformation. Mol Plant 7:1793–1796
Pham AT, Lee JD, Shannon JG, Bilyeu KD (2010) Mutant alleles of FAD2-1A and FAD2-1B combine to produce soybeans with the high oleic acid seed oil trait. BMC Plant Biol 10:195
Purkayastha A, Dasgupta I (2009) Virus-induced gene silencing: a versatile tool for discovery of gene functions in plants. Plant Physiol Biochem 47:967–976
Ramegowda V, Mysore KS, SenthilKumar M (2014) Virus-induced gene silencing is a versatile tool for unraveling the functional relevance of multiple abiotic-stress-responsive genes in crop plants. Front Plant Sci. doi:10.3389/fpls.2014.00323
Sashital D, Doudna JA (2010) Structural insights into RNA interference. Curr Opin Struct Biol 20:90–97
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8:517–527
Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18:1121–1133
Senthil-Kumar M, Mysore KS (2011) Caveat of RNAi in plants: the off-target effect. Methods Mol Biol 744:13–25
Travella S, Klimm TE, Keller B (2006) RNA interference-based gene silencing as an efficient tool for functional genomics in hexaploid bread wheat. Plant Physiol 142:6–20
Voinnet O (2008) Use, tolerance and avoidance of amplified RNA silencing by plants. Trends Plant Sci 3:317–328
Watson JM, Fusaro AF, Wang M, Waterhouse PM (2005) RNA silencing platforms in plants. FEBS Lett 579:5982–5987
Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q et al (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27:581–590
Zamore P, Tuschl T, Sharp P, Bartel D (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101:25–33
Zeng P, Vadnais D, Zhang Z, Polacco J (2004) Refined glufosinate selection in Agrobacterium-mediated transformation of soybean [Glycine max (L.) Merr.]. Plant Cell Rep 22:478–482
Zhang Z (2014) Artificial trans-acting small interfering RNA: a tool for plant biology study and crop improvements. Planta 239:1139–1146
Acknowledgments
We thank Dr. David Setzer for allowing us to use his laboratory and the equipment for primer extension analysis. Thanks are also expended to: Dr. Joann R. De Tar for her technical assistance in Glycine max FAD2-1B atasiRNA design, and Mr. Neng Wang for his Arabidopsis planting and plant care as well as Dr. William Folk and Mrs. Theresa Musket for proof-reading. This work was supported by the Mid-America Research and Development Foundation and the Missouri Soybean Merchandising Council Grant (Project No. 11-338).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Z.-Y. Wang.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Baykal, U., Liu, H., Chen, X. et al. Novel constructs for efficient cloning of sRNA-encoding DNA and uniform silencing of plant genes employing artificial trans-acting small interfering RNA. Plant Cell Rep 35, 2137–2150 (2016). https://doi.org/10.1007/s00299-016-2024-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-016-2024-9