Abstract
Key message
A FRUITFULL homolog GmFULa was cloned and found to play roles in the flowering and maturation of soybean.
Abstract
Soybean varieties exhibit great diversity in terms of flowering and maturation due to differences in their photoperiodic responses. The underlying mechanism remains unclear despite the fact that some upstream flowering genes have been studied. FRUITFULL (FUL) genes are one group of downstream flowering genes known to have major roles in reproductive transition, floral meristem identity, and floral organ identity. However, FUL homologs and their functions are poorly understood in soybean. Here, a soybean FUL homolog was cloned from the late-maturing photoperiod-sensitive soybean variety Zigongdongdou (ZGDD) and designated GmFULa. In ZGDD, GmFULa exhibited a terminal-preferential expression pattern, with higher expression in the root and shoot apices than in the middle parts. Diurnal rhythm analysis revealed that photoperiod regulates the GmFULa expression level but does not alter its diurnal rhythm. ZGDD was maintained under different photoperiod conditions (long day, LD; short day, SD; LD after 13 short days, SD13-LD) to assess GmFULa expression in newly expanded leaves and in the shoot apex. From this analysis, GmFULa expression was detected in the floral meristem, floral organs and their primordia; trifoliate leaves; and the inflorescence meristem, with the expression levels induced by SD and inhibited by LD. GmFULa expression was also associated with maturity in seven soybean varieties with different photoperiod sensitivities. Therefore, photoperiod conditions affect the expression level of GmFULa but not its diurnal rhythm. The gene plays pleiotropic roles in reproductive transition, flowering, and leaf development and is associated with maturity in soybean.









Similar content being viewed by others
Abbreviations
- FUL:
-
FRUITFULL
- CDS:
-
Coding sequence
- LD:
-
Long day
- SD:
-
Short day
- AP1:
-
APETALA1
- ZGDD:
-
Zigongdongdou
- HH27:
-
Heihe 27
References
Bemer M, Karlova R, Ballester AR, Tikunov YM, Bovy AG, Wolters-Arts M, Rossetto PB, Angenent GC, de Maagd RA (2012) The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 24:4437–4451
Benlloch R, D’Erfurth I, Ferrandiz C, Cosson V, Beltran JP, Canas LA, Kondorosi A, Madueno F, Ratet P (2006) Isolation of mtpim proves Tnt1 a useful reverse genetics tool in Medicago truncatula and uncovers new aspects of AP1-like functions in legumes. Plant Physiol 142:972–983
Berbel A, Navarro C, Ferrandiz C, Canas LA, Madueno F, Beltran JP (2001) Analysis of PEAM4, the pea AP1 functional homologue, supports a model for AP1-like genes controlling both floral meristem and floral organ identity in different plant species. Plant J 25:441–451
Berbel A, Ferrandiz C, Hecht V, Dalmais M, Lund OS, Sussmilch FC, Taylor SA, Bendahmane A, Ellis TH, Beltran JP, Weller JL, Madueno F (2012) VEGETATIVE1 is essential for development of the compound inflorescence in pea. Nat Commun 3:797
Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119:721–743
Burko Y, Shleizer-Burko S, Yanai O, Shwartz I, Zelnik ID, Jacob-Hirsch J, Kela I, Eshed-Williams L, Ori N (2013) A role for APETALA1/FRUITFULL transcription factors in tomato leaf development. Plant Cell 25:2070–2083
Chen D, Guo B, Hexige S, Zhang T, Shen D, Ming F (2007) SQUA-like genes in the orchid Phalaenopsis are expressed in both vegetative and reproductive tissues. Planta 226:369–380
Chi Y, Huang F, Liu H, Yang S, Yu D (2011) An APETALA1-like gene of soybean regulates flowering time and specifies floral organs. J Plant Physiol 168:2251–2259
Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316:1030–1033
Danilevskaya ON, Meng X, Selinger DA, Deschamps S, Hermon P, Vansant G, Gupta R, Ananiev EV, Muszynski MG (2008) Involvement of the MADS-box gene ZMM4 in floral induction and inflorescence development in maize. Plant Physiol 147:2054–2069
Dreni L, Kater MM (2013) MADS reloaded: evolution of the AGAMOUS subfamily genes. New Phytol
Fei Z, Jia Z, Leng J, Zhang B, Wu C (2009) Identification of photoperiodic responses of different soybean ecotypes. Crops, pp 46–49
Fornara F, Parenicova L, Falasca G, Pelucchi N, Masiero S, Ciannamea S, Lopez-Dee Z, Altamura MM, Colombo L, Kater MM (2004) Functional characterization of OsMADS18, a member of the AP1/SQUA subfamily of MADS box genes. Plant Physiol 135:2207–2219
Gocal GF, King RW, Blundell CA, Schwartz OM, Andersen CH, Weigel D (2001) Evolution of floral meristem identity genes. Analysis of Lolium temulentum genes related to APETALA1 and LEAFY of Arabidopsis. Plant Physiol 125:1788–1801
Gu Q, Ferrandiz C, Yanofsky MF, Martienssen R (1998) The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125:1509–1517
Hao G, Chen X, Pu M (1992) Maturity groups of soybean cultivars in China. Acta Agron Sin 18:275–281
Hartwig EE (1970) Growth and reproductive characteristics of soybeans (Glycine max (L.) Merr.) grown under short-day conditions. Trop Sci 12:47–53
Huang G, Ma J, Han Y, Chen X, Fu Y (2011) Cloning and expression analysis of the soybean CO-like gene GmCOL9. Plant Mol Biol Rep 29:352–359
Jaakola L, Poole M, Jones MO, Kamarainen-Karppinen T, Koskimaki JJ, Hohtola A, Haggman H, Fraser PD, Manning K, King GJ, Thomson H, Seymour GB (2010) A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol 153:1619–1629
Jia Z, Wu CX, Wang M, Sun HB, Hou WS, Jiang BJ, Han TF (2011) Effects of leaf number of stock or scion in graft union on scion growth and development of soybean. Acta Agron Sin, pp 650–660
Jian B, Liu B, Bi Y, Hou W, Wu C, Han T (2008) Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol Biol 9:59
Jiang Y, Han YZ, Zhang XM (2011) Expression profiles of a CONSTANS homologue GmCOL11 in Glycine max. Russ J Plant Physiol 58:928–935
Jiang B, Yue Y, Gao Y, Ma L, Sun S, Wu C, Hou W, Lam H, Han T (2013) GmFT2a polymorphism and maturity diversity in soybeans. PLoS One 8:e77474
Jung J, Ju Y, Seo PJ, Lee J, Park C (2012) The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J 69:577–588
Kong F, Liu B, Xia Z, Sato S, Kim B, Watanabe S, Yamada T, Tabata S, Kanazawa A, Harada K, Abe J (2010) Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol 154(3):1220–1231
Kramer EM, Hall JC (2005) Evolutionary dynamics of genes controlling floral development. Curr Opin Plant Biol 8:13–18
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Litt A (2007) An evaluation of A-function: evidence from the APETALA1 and APETALA2 gene lineages. Int J Plant Sci 168:73–91
Litt A, Irish VF (2003) Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: implications for the evolution of floral development. Genetics 165:821–833
Liu B, Kanazawa A, Matsumura H, Takahashi R, Harada K, Abe J (2008) Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 180:995–1007
Liu B, Watanabe S, Uchiyama T, Kong F, Kanazawa A, Xia Z, Nagamatsu A, Arai M, Yamada T, Kitamura K, Masuta C, Harada K, Abe J (2010) The soybean stem growth habit gene Dt1 is an ortholog of Arabidopsis TERMINAL FLOWER1. Plant Physiol 153:198–210
Liu L, Ma J, Han Y, Chen X, Fu YF (2011) The isolation and analysis of a soybean CO homologue GmCOL10. Russ J Plant Physiol 58:330–336
Mandel MA, Yanofsky MF (1995) The Arabidopsis AGL8 MADS box gene is expressed in inflorescence meristems and is negatively regulated by APETALA1. Plant Cell 7:1763–1771
Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360:273–277
McGarry RC, Kragler F (2013) Phloem-mobile signals affecting flowers: applications for crop breeding. Trends Plant Sci 18:198–206
Melzer S, Lens F, Gennen J, Vanneste S, Rohde A, Beeckman T (2008) Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat Genet 40:1489–1492
Muller BM, Saedler H, Zachgo S (2001) The MADS-box gene DEFH28 from Antirrhinum is involved in the regulation of floral meristem identity and fruit development. Plant J 28:169–179
Pabon-Mora N, Ambrose BA, Litt A (2012) Poppy APETALA1/FRUITFULL orthologs control flowering time, branching, perianth identity, and fruit development. Plant Physiol 158:1685–1704
Preston JC, Kellogg EA (2008) Discrete developmental roles for temperate cereal grass VERNALIZATION1/FRUITFULL-like genes in flowering competency and the transition to flowering. Plant Physiol 146:265–276
Sather DN, Golenberg EM (2009) Duplication of AP1 within the Spinacia oleracea L. AP1/FUL clade is followed by rapid amino acid and regulatory evolution. Planta 229:507–521
Sun H, Jia Z, Cao D, Jiang B, Wu C, Hou W, Liu Y, Fei Z, Zhao D, Han T (2011) GmFT2a, a soybean homolog of FLOWERING LOCUS T, is involved in flowering transition and maintenance. PLoS One 6:e29238
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Wang G (1981) The research about the ecological division of soybean cultivars in China. Sci Agri Sin 39–46
Watanabe S, Hideshima R, Xia Z, Tsubokura Y, Sato S, Nakamoto Y, Yamanaka N, Takahashi R, Ishimoto M, Anai T, Tabata S, Harada K (2009) Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics 182:1251–1262
Watanabe S, Xia Z, Hideshima R, Tsubokura Y, Sato S, Yamanaka N, Takahashi R, Anai T, Tabata S, Kitamura K, Harada K (2011) A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 188:260–395
Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309:1056–1059
Wong CE, Singh MB, Bhalla PL (2013) Novel members of the AGAMOUS LIKE 6 subfamily of MIKCC-type MADS-box genes in soybean. BMC Plant Biol 13:105
Wu C, Ma Q, Yam KM, Cheung MY, Xu Y, Han T, Lam HM, Chong K (2006) In situ expression of the GmNMH7 gene is photoperiod-dependent in a unique soybean [Glycine max (L.) Merr.] flowering reversion system. Planta 223:725–735
Xia Z, Watanabe S, Yamada T, Tsubokura Y, Nakashima H, Zhai H, Anai T, Sato S, Yamazaki T, Lü S, Wu H, Tabata S, Harada K (2012) Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc Natl Acad Sci USA 109:E2155–E2164
Xu YY, Chong K, Xu ZH, Tan KH (2002) The practical technique of in situ hybridization with RNA probe. Chin Bull Bot 19:234–238
Yu H, Xu Y, Tan EL, Kumar PP (2002) AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proc Natl Acad Sci USA 99:16336–16341
Zhang LX, Kyei-Boahen S, Zhang J, Zhang MH, Freeland TB, Watson Jr CE, Liu XM (2007) Modifications of optimum adaptation zones for soybean maturity groups in the USA. Crop Manag. doi:10.1094/CM-2007-0927-01-RS
Zhang Q, Ma J, Chen X, Fu Y (2010) Cloning and analysis of GmCOL4 gene in Glycine max. Acta Agron Sin 36:539–548
Zhang W, Fan S, Pang C, Wei H, Ma J, Song M, Yu S (2013) Molecular cloning and function analysis of two SQUAMOSA-like MADS-box genes from Gossypium hirsutum L. J Integr Plant Biol 55(7):597–607
Acknowledgments
This work was supported by the China Agriculture Research System (CARS-04), the National Natural Science Foundation of China (http://www.nsfc.gov.cn; Grant No. 30070456 and 30471054) and the Chinese Academy of Agricultural Sciences (CAAS) Innovation Project.
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Renate Schmidt.
Z. Jia and B. Jiang contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
299_2014_1693_MOESM1_ESM.tif
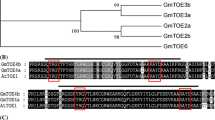
Additional file 1 - Figure S1. GmFULa DNA and protein sequences. Non-bold, GmFULa DNA sequence; bold, GmFULa protein sequence; yellow, GmFULa coding sequence; red, the conserved C-terminal motif of euFUL and FUL-like proteins. (TIFF 1016 kb)
299_2014_1693_MOESM4_ESM.tif
Additional file 4 - Figure S4. GmFULa expression in the shoot apices of different soybean varieties under short-day conditions by in situ hybridization. (a)-(c) Shoot apices of soybean variety Heihe 27. (d)-(f) Shoot apices of soybean variety Edou 4. (g)-(i) Shoot apices of soybean variety Jilin 30. (j)-(l) Shoot apices of soybean variety Jindou 19. (m)-(o) Shoot apices of soybean variety Dian 86-5. (p)-(r) Shoot apices of soybean variety Huaxia 1. SD5, SD10, and SD15 indicate 5, 10, and 15 short days, respectively. am, apical meristem; axm, axillary meristem; br, bract; c, carpel; fm, floral meristem; fp, floral primordium; im, inflorescence meristem; ip, inflorescence primordium; le, leaf/trifoliate leaf; p, pistil; pe, petal; pep, petal primordium; pp, pistil primordium; rf, reversed flower; s, stamen; se, sepal; sep, sepal primordium; sp, stamen primordium; tlp, trifoliate leaf primordium. Bar, 250 μM (TIFF 10444 kb)
Rights and permissions
About this article
Cite this article
Jia, Z., Jiang, B., Gao, X. et al. GmFULa, a FRUITFULL homolog, functions in the flowering and maturation of soybean. Plant Cell Rep 34, 121–132 (2015). https://doi.org/10.1007/s00299-014-1693-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-014-1693-5