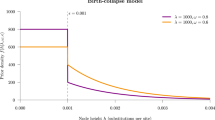
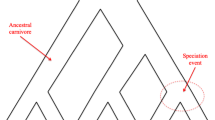
Abstract
The focus of this article is a Bayesian method for inferring both species delimitations and species trees under the multispecies coalescent model using molecular sequences from multiple loci. The species delimitation requires no a priori assignment of individuals to species, and no guide tree. The method is implemented in a package called STACEY for BEAST2, and is a extension of the author’s DISSECT package. Here we demonstrate considerable efficiency improvements by using three new operators for sampling from the posterior using the Markov chain Monte Carlo algorithm, and by using a model for the population size parameters along the branches of the species tree which allows these parameters to be integrated out. The correctness of the moves is demonstrated by tests of the implementation. The practice of using a pipeline approach to species delimitation under the multispecies coalescent, has been shown to have major problems on simulated data (Olave et al. in Syst Biol 63:263–271. doi:10.1093/sysbio/syt106, 2014). The same simulated data set is used to demonstrate the accuracy and improved convergence of the present method. We also compare performance with *BEAST for a fixed delimitation analysis on a large data set, and again show improved convergence.







Similar content being viewed by others
References
Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ (2014) BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10(4):e1003,537. doi:10.1371/journal.pcbi.1003537
Degnan JH, Rosenberg NA (2009) Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol Evol 24:332–340
Edwards SV (2009) Is a new and general theory of molecular systematics emerging? Evolution 63:1–19
Felsenstein J (2003) Inferring phylogenies. Sinauer Associates, Sunderland. doi:10.1016/S0022-0000(02)00003-X
Flot JF (2015) Species delimitation’s coming of age. Syst Biol 64(6):897–899
Giarla T, Esselstyn J (2015) The challenges of resolving a rapid, recent radiation: empirical and simulated phylogenomics of Philippine shrews. Syst Biol 64(5):727–740. doi:10.1093/sysbio/syv029
Heled J, Drummond A (2010) Bayesian inference of species trees from multilocus data. Mol Biol Evol 27:570–580
Hey J, Nielsen R (2007) Integration within the felsenstein equation for improved markov chain Monte Carlo methods in population genetics. Proc Natl Acad Sci 104:2785–2790
Höhna S, Defoin-Platel M, Drummond AJ (2008) Clock-constrained tree proposal operators in Bayesian phylogenetic inference. In: 8th IEEE international conference on bioinformatics and bioengineering, Athens, Greece, pp 1–7, 8–10 Oct 2008
Huang H, He Q, Kubatko LS, Knowles LL (2010) Sources of error for species-tree estimation: impact of mutational and coalescent effects on accuracy and implications for choosing among different methods. Syst Biol 59:573–583
Huelsenbeck JP, Andolfatto P (2007) Inference of population structure under a Dirichlet process model. Genetics 175:1787–1802
Jones G, Aydin Z, Oxelman B (2014) DISSECT: an assignment-free Bayesian discovery method for species delimitation under the multispecies coalescent. Bioinformatics. doi:10.1093/bioinformatics/btu770
Liu L, Pearl DK, Brumfield RT, Edwards SV (2008) Estimating species trees using multiple allele DNA sequence data. Evolution 62(8):2080–2091
Olave M, Solà E, Knowles LL (2014) Upstream analyses create problems with DNA-based species delimitation. Syst Biol 63:263–271. doi:10.1093/sysbio/syt106
Plummer M, Best N, Cowles K, Vines K (2006) CODA: convergence diagnosis and output analysis for MCMC. R News 6(1), 7–11. http://CRAN.R-project.org/doc/Rnews/
Pritchard JK, Stephens M, Donnelly PJ (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rannala B (2015) The art and science of species delimitation. Curr Zool 61:846–853
Rannala B, Yang Z (2003) Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics 164:1645–1656
Rannala B, Yang Z (2013) Improved reversible jump algorithms for Bayesian species delimitation. Genetics 194:245–253
Solís-Lemus C, Knowles LL, Ane C (2015) Bayesian species delimitation combining multiple genes and traits in a unified framework. Evolution 69:492–507
Yang Z (2002) Likelihood and Bayes estimation of ancestral population sizes in hominoids using data from multiple loci. Genetics 162:1811–1823
Yang Z, Rannala B (2010) Bayesian species delimitation using multilocus sequence data. Proc Natl Acad Sci USA 107:9264–9269
Yang Z, Rannala B (2014) Unguided species delimitation using DNA sequence data from multiple loci. Mol Biol Evol 31(12):3125–3135. doi:10.1093/molbev/msu279
Zhang C, Rannala B, Yang Z (2014) Bayesian species delimitation can be robust to guide-tree inference errors. Syst Biol 63:993–1004. doi:10.1093/sysbio/syu052
Acknowledgments
I thank the developers of BEAST for making this work feasible, and Remco Bouckaert in particular for helpful advice on writing the STACEY package. I thank the authors of Olave et al. (2014) and Giarla and Esselstyn (2015) for making their simulated data readily available, and for supplying extra details about their simulations. I thank two anonymous reviewers for valuable comments on an earlier version of this paper.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jones, G. Algorithmic improvements to species delimitation and phylogeny estimation under the multispecies coalescent. J. Math. Biol. 74, 447–467 (2017). https://doi.org/10.1007/s00285-016-1034-0
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-016-1034-0