Abstract
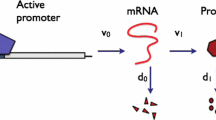
Stochastic models for gene expression frequently exhibit dynamics on several different scales. One potential time-scale separation is caused by significant differences in the lifetimes of mRNA and protein; the ratio of the two degradation rates gives a natural small parameter in the resulting chemical master equation, allowing for the application of perturbation techniques. Here, we develop a framework for the analysis of a family of ‘fast-slow’ models for gene expression that is based on geometric singular perturbation theory. We illustrate our approach by giving a complete characterisation of a standard two-stage model which assumes transcription, translation, and degradation to be first-order reactions. In particular, we present a systematic expansion procedure for the probability-generating function that can in principle be taken to any order in the perturbation parameter, allowing for an approximation of the corresponding propagator probabilities to that same order. For illustrative purposes, we perform this expansion explicitly to first order, both on the fast and the slow time-scales; then, we combine the resulting asymptotics into a composite fast-slow expansion that is uniformly valid in time. In the process, we extend, and prove rigorously, results previously obtained by Shahrezaei and Swain (Proc Natl Acad Sci USA 105(45):17256–17261, 2008) and Bokes et al. (J Math Biol 64(5):829–854, 2012; J Math Biol 65(3):493–520, 2012). We verify our asymptotics by numerical simulation, and we explore its practical applicability and the effects of a variation in the system parameters and the time-scale separation. Focussing on biologically relevant parameter regimes that induce translational bursting, as well as those in which mRNA is frequently transcribed, we find that the first-order correction can significantly improve the steady-state probability distribution. Similarly, in the time-dependent scenario, inclusion of the first-order fast asymptotics results in a uniform approximation for the propagator probabilities that is superior to the slow dynamics alone. Finally, we discuss the generalisation of our geometric framework to models for regulated gene expression that involve additional stages.






Similar content being viewed by others
References
Amann H (1990) Ordinary differential equations. An introduction to nonlinear analysis, translated from the German by Gerhard Metzen. In: de Gruyter studies in mathematics, vol 13. Walter de Gruyter & Co., Berlin
Arrowsmith DK, Place CM (1990) An introduction to dynamical systems. Cambridge University Press, Cambridge
Abramowitz M, Stegun IA (eds) (1972) Handbook of mathematical functions with formulas, graphs, and mathematical tables, national bureau of standards. Applied mathematics series, vol 55. Dover Publications, New York
Balser W (1994) From divergent power series to analytic functions. In: Theory and application of multisummable power series. Lecture notes in mathematics, vol 1582. Springer, Berlin
Bennett MR, Hasty J (2009) Microfluidic devices for measuring gene network dynamics in single cells. Nat Rev Genet 10(9):628–638
Bernstein J, Khodursky A, Lin P, Lin-Chao S, Cohen S (2002) Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc Natl Acad Sci USA 99(15):9697–9702
Blasi T, Buettner F, Strasser M, Linnarsson S, Marr C, Theis FJ (2013) Cell-cycle effects obscure true kinetics of gene expression in single cell transcriptomics data. (in preparation)
Bokes P, King JR, Wood ATA, Loose M (2012) Exact and approximate distributions of protein and mRNA levels in the low-copy regime of gene expression. J Math Biol 64(5):829–854
Bokes P, King JR, Wood ATA, Loose M (2012) Multiscale stochastic modelling of gene expression. J Math Biol 65(3):493–520
Bowsher CG, Swain PS (2012) Identifying sources of variation and the flow of information in biochemical networks. Proc Natl Acad Sci USA 109(20):E1320–E1328
Cao Y, Gillespie DT, Petzold LR (2005) The slow-scale stochastic simulation algorithm. J Chem Phys 122(1):014116
Canalis-Durand M, Ramis JP, Schäfke R, Sibuya Y (2000) Gevrey solutions of singularly perturbed differential equations. J Reine Angew Math 518:95–129
Cotter SL, Zygalakis KC, Kevrekidis IG, Erban R (2011) A constrained approach to multiscale stochastic simulation of chemically reacting systems. J Chem Phys 135(9):094102
Cover TM, Thomas JA (2006) Elements of information theory. In: Wiley series in telecommunications and signal processing, 2nd edn. Wiley, Hoboken
Dumortier F, Roussarie R (1996) Canard cycles and center manifolds. Mem Am Math Soc 121(577)
Erban R, Kevrekidis IG, Adalsteinsson D, Elston TC (2006) Gene regulatory networks: a coarse-grained, equation-free approach to multiscale computation. J Chem Phys 124(8):084106
Fenichel N (1979) Geometric singular perturbation theory for ordinary differential equations. J Differ Equ 31(1):53–98
Friedman N, Cai L, Xie XS (2006) Linking stochastic dynamics to population distribution: an analytical framework of gene expression. Phys Rev Lett 97(16):168302
García-Martínezm J, González-Candelas F, Pérez-Ortín J (2007) Common gene expression strategies revealed by genome-wide analysis in yeast. Genome Biol 8:R222
Gillespie DT (1977) Exact stochastic simulation of coupled chemical reactions. J Phys Chem 81(25):2340–2361
Gillespie DT (1992) A rigorous derivation of the chemical master equation. Phys A 188(1–3):404–425
Gillespie DT (2009) Deterministic limit of stochastic chemical kinetics. J Phys Chem 113(6):1640–1644
Golding I, Paulsson J, Zawilski SM, Cox EC (2005) Real-time kinetics of gene activity in individual bacteria. Cell 123(6):1025–1036
Grima R (2011) Construction and accuracy of partial differential equation approximations to the chemical master equation. Phys Rev E 84(5):056109
Grima R, Schmidt D, Newman TJ (2012) Steady-state fluctuations of a genetic feedback loop: an exact solution. J Chem Phys 137(3):035104
Harper CV, Finkenstädt B, Woodcock DJ, Friedrichsen S, Semprini S, Ashall L, Spiller DG, Mullins JJ, Rand DA, Davis JRE, White MRH (2011) Dynamic analysis of stochastic transcription cycles. PLoS Biol 9(4):e1000607
Hek G (2010) Geometric singular perturbation theory in biological practice. J Math Biol 60(3):347–386
Jahnke T, Huisinga W (2007) Solving the chemical master equation for monomolecular systems analytically. J Math Biol 54(1):1–26
Jones CKRT (1995) Geometric singular perturbation theory. In Dynamical systems (Montecatini Terme, 1994). Lecture notes in Math., vol 1609. Springer, Berlin, pp 44–118
Kaern M, Elston TC, Blake WJ, Collins JJ (2005) Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet 6(6):451–464
Krupa M, Szmolyan P (2001) Extending geometric singular perturbation theory to non-hyperbolic points—fold and canard points in two dimensions. SIAM J Math Anal 33(2):286–314
Lagerstrom PA (2010) Matched asymptotic expansions: ideas and techniques. In: Applied mathematical sciences, vol 76. Springer, New York
Laurenzi IJ (2000) An analytical solution of the stochastic master equation for reversible bimolecular reaction kinetics. J Chem Phys 113(8):3315–3322
Locke JC, Elowitz MB (2009) Using movies to analyse gene circuit dynamics in single cells. Nat Rev Microbiol 7(5):383–392
Mansour MM, Van den Broeck C, Nicolis G, Turner JW (1981) Asymptotic properties of markovian master equations. Ann Phys 131(2):283–313
Mastny EA, Haseltine EL, Rawlings JB (2007) Two classes of quasi-steady-state model reductions for stochastic kinetics. J Chem Phys 127(9):094106
McAdams H, Arkin A (1997) Stochastic mechanisms in gene expression. Proc Natl Acad Sci USA 94(3):814–819
Mukhopadhyay P (2011) An introduction to the theory of probability. World Scientific Publishing Co., Singapore
Murray JD (2008) Mathematical biology I: an introduction. In: Interdisciplinary applied mathematics, vol 17, 3rd edn. Springer, New York
Peccoud J, Ycart B (1995) Markovian modeling of gene-product synthesis. Theor Popul Biol 48(2):222–234
Popović N, Marr C, Swain PS (2015) A dynamical systems framework for parameter inference in stochastic gene expression. (in preparation)
Pendar H, Platini T, Kulkarni RV (2013) Exact protein distributions for stochastic models of gene expression using partitioning of Poisson processes. Phys Rev E 87(4):042720
Raser JM, O’Shea EK (2004) Control of stochasticity in eukaryotic gene expression. Science 304(5678):1811–1814
Risken H (1989) The Fokker–Planck equation: methods of solution and applications. In: Springer series in synergetics, vol 18, 2nd edn. Springer, Berlin
Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M (2011) Global quantification of mammalian gene expression control. Nature 473(7347):337–342 [Corrigendum: Nature 495(7439), 126–127, 2013]
Segel LA, Slemrod M (1989) The quasi-steady-state assumption: a case study in perturbation. SIAM Rev 31(3):446–477
Strogatz SH (1994) Nonlinear dynamics and chaos: with applications to physics, biology, chemistry, and engineering. In: Studies in nonlinearity. Addison-Wesley Publishing Company, Massachusetts
Swain P, Elowitz MB, Siggia ED (2002) Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA 99(20):12795–12800
Shahrezaei V, Swain PS (2008) The stochastic nature of biochemical networks. Curr Opin Biotechnol 19:369–374
Shahrezaei V, Swain PS (2008) Analytical distributions for stochastic gene expression. Proc Natl Acad Sci USA 105(45):17256–17261
Sanft KR, Wu S, Roh M, Fu J, Lim RK, Petzold LR (2011) StochKit2: software for discrete stochastic simulation of biochemical systems with events. Bioinformatics 27(17):2457–2458
Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F (2011) Mammalian genes are transcribed with widely different bursting kinetics. Science 332(6028):472–474
Thattai M, van Oudenaarden A (2001) Intrinsic noise in gene regulatory networks. Proc Natl Acad Sci USA 98(15):151588–151598
Thomas P, Straube AV, Grima R (2012) The slow-scale linear noise approximation: an accurate, reduced stochastic description of biochemical networks under timescale separation conditions. BMC Syst Biol 6(1):39
Verhulst F (2005) Methods and applications of singular perturbations: boundary layers and multiple timescale dynamics. In: Texts in applied mathematics, vol 50. Springer, New York
Viterbi A (1967) Error bounds for convolutional codes and an asymptotically optimum decoding algorithm. IEEE Trans Inf Theory 13(2):260–269
Wang Y, Liu C, Storey J, Tibshirani R, Herschlag D, Brown P (2002) Precision and functional specificity in mRNA decay. Proc Natl Acad Sci USA 99(9):5860–5865
Weinan E, Liu D, Vanden-Eijnden E (2005) Nested stochastic simulation algorithm for chemical kinetic systems with disparate rates. J Chem Phys 123(19):194107
Wiggins S (2003) Introduction to applied nonlinear dynamical systems and chaos. In: Texts in applied mathematics, 2nd edn. Springer, New York
Yu J, Xiao J, Ren X, Lao K, Xie X (2006) Probing gene expression in live cells, one protein molecule at a time. Science 311(5767):1600–1603
Zauderer E (2006) Partial differential equations of applied mathematics. In: Pure and applied mathematics (New York), 3rd edn. Wiley, Hoboken
Zopf CJ, Quinn K, Zeidman J, Maheshri N (2013) Cell-cycle dependence of transcription dominates noise in gene expression. PLoS Comput Biol 9(7):1–12
Author information
Authors and Affiliations
Corresponding author
Additional information
Nikola Popović is grateful to Peter De Maesschalck and Ramon Grima for their careful reading of drafts of the manuscript and numerous helpful suggestions, as well as to Peter Szmolyan for stimulating discussions. Moreover, the authors acknowledge grant support from the Moray Endowment Fund, as well as from MAXIMATHS, an initiative by the School of Mathematics at the University of Edinburgh aimed at maximising the impact of mathematics in science and engineering. Finally, the authors thank three anonymous referees for valuable comments which greatly improved the original manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Popović, N., Marr, C. & Swain, P.S. A geometric analysis of fast-slow models for stochastic gene expression. J. Math. Biol. 72, 87–122 (2016). https://doi.org/10.1007/s00285-015-0876-1
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-015-0876-1
Keywords
- Stochastic gene expression
- Chemical master equation
- Two-stage model
- Generating function
- Propagator probabilities
- Asymptotic expansion
- Geometric singular perturbation theory