Abstract
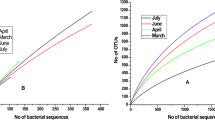
Mine water is an example of an extreme environment that contains a large number of diverse and specific bacteria. It is imperative to gain an understanding of these bacterial communities in order to develop effective strategies for the bioremediation of polluted aquatic systems. In this study, the high-throughput sequencing approach was used to characterize the bacterial communities in two different mine waters of South Africa: vanadium and gold mine water. Over 2629 operational taxonomic units (OTUs) were recovered from 15,802 reads of the 16S ribosomal RNA (rRNA) gene. They represented 8 phyla, 43 orders, 84 families and 105 genera. Proteobacteria and unclassified bacterial sequences were the most dominant. Apart from these, Firmicutes, Bacteroidetes, Actinobacteria, Candidate phylum OD1, Cyanobacteria, Verrucomicrobia and Deinococcus-Thermus were the recovered phyla, although their relative abundance differed between both the mine-water samples. Yet, diversity indices suggested that the bacterial communities inhabiting the vanadium mine water were more diverse than those in gold mine water. Interestingly, substantial percentages of the reads from either sample (58 % in vanadium and 17 % in gold mine water) could not be assigned to any phylum and remained unclassified, suggesting hitherto unidentified populations, and vast untapped microbial diversity. Overall, the results of this study exhibited bacterial community structures with high diversity in mine water, which can be explored further for their role in bioremediation and environmental management.



Similar content being viewed by others
References
Adler RA, Claassen M, Godfrey L, Turton AR (2007) Water, mining, and waste: an historical and economic perspective on conflict management in South Africa. Econ Peace Secur J 2:33–44
An C, Kuda T, Yazaki T, Takahashi H, Kimura B (2014) Caecal environment of rats fed far East Asian-modelled diets. Appl Microbiol Biotechnol. doi:10.1007/s00253-014-5535-5538
APHA (2001) Standard methods for the examination of water and wastewater, 20th edn. American Public Health Association (APHA), Washington DC
Aslam Z, Yasir M, Khaliq A, Matsui K, Chung YR (2010) Too much bacteria still unculturable. Crop Environ 1:59–60
Berg J, Brandt KK, Al-Soud WA, Holm PE, Hansen LH, Sørensen SJ, Nybroe O (2012) Long-term Cu exposure selects for Cu-tolerant bacterial communities with altered composition, but unaltered richness. Appl Environ Microbiol 78:7438–7446
Bohorquez LC, Delgado-Serrano L, López G, Osorio-Forero C, Klepac-Ceraj V, Kolter R, Junca H, Baena S, Zambrano MM (2012) In-depth characterization via complementing culture-independent approaches of the microbial community in an acidic hot spring of the Colombian Andes. Microb Ecol 63:103–115
Bokulich NA, Mills DA (2013) Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl Environ Microbiol 79:2519–2526
Bond PL, Smriga SP, Banfield JF (2000) Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl Environ Microbiol 66:3842–3849
Brazelton WJ, Ludwig KA, Sogin ML, Andreishcheva EN, Kelley DS, Shen C-C, Edwards RL, Baross JA (2010) Archaea and bacteria with surprising microdiversity show shifts in dominance over 1,000-year time scales in hydrothermal chimneys. Proc Natl Acad Sci U S A 107:1612–1617
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624
Dopson M, Baker-Austin C, Koppineedi PR, Bond PL (2003) Growth in sulfidic mineral environments: metal resistance mechanisms in acidophilic micro-organisms. Microbiology 149:1959–1970
Druschel GK, Baker BJ, Gihring TM, Banfield JF (2004) Acid mine drainage biogeochemistry at Iron Mountain, California. Geochem Trans 5:13–32
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Gansauge M-T, Meyer M (2013) Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat Protoc 8:737–748
Garcia-Moyano A, Gonzalez-Toril E, Aguilera A, Amils R (2007) Prokaryotic community composition and ecology of floating macroscopic filaments from an extreme acidic environment, Rio Tinto (SW, Spain). Syst Appl Microbiol 30:601–614
Goebel BM, Stackebrandt E (1994) Cultural and phylogenetic analysis of mixed microbial populations found in natural and commercial bioleaching environments. Appl Environ Microbiol 60:1614–1621
Gołębiewski M, Deja-Sikora E, Cichosz M, Tretyn A, Wróbel B (2014) 16S rDNA pyrosequencing analysis of bacterial community in heavy metals polluted soils. Microb Ecol 67:635–647
Gonzalez-Toril E, Llobet-Brossa E, Casamayor EO, Amann R, Amils R (2003) Microbial ecology of an extreme acidic environment, the Tinto River. Appl Environ Microbiol 69:4853–4865
Gough HL, Stahl DA (2011) Microbial community structures in anoxic freshwater lake sediment along a metal contamination gradient. ISME J 5:543–558
He Z, Xie X, Xiao S, Li J, Qiu G (2007) Microbial diversity of mine water at Zhong Tiaoshan copper mine, China. J Basic Microbiol 47:485–495
Hennecke H, Kjelleberg S, Brussaard C (2013) Molecular insights into environmental microbes. Fems Microbiol Rev 37:285
Hugenholtz P, Goebel BM, Pace NR (1998) Impact of culture independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol 180:4765–4774
Imarla T, Hector SB, Deane SM, Rawlings DE (2006) Resistance determinants of a highly arsenic-resistant strain of Leptospirillum ferriphilum isolated from a commercial biooxidation tank. Appl Environ Microbiol 72:2247–2253
Johnson DB, Hallberg KB (2003) The microbiology of acidic mine waters. Res Microbiol 154:466–473
Kamika I, Momba MNB (2013) Microbial diversity of Emalahleni mine water in South Africa and tolerance ability of the predominant organism to vanadium and nickel. Plos One 9:e86189
Kuang J-L, Huang L-N, Chen L-X, Hua Z-S, Li S-J, Hu M, Li J-T, Shu W-S (2013) Contemporary environmental variation determines microbial diversity patterns in acid mine drainage. ISME J 7:1038–1050
Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, Knight R (2012) Experimental and analytical tools for studying the human microbiome. Nat Rev Genet 13:47–58
Kyrpides NC, Hugenholtz P, Eisen JA, Woyke T, Göker M, Parker CT, Amann R, Beck BJ, Chain PS, Chun J, Colwell RR, Danchin A, Dawyndt P, Dedeurwaerdere T, DeLong EF, Detter JC, De Vos P, Donohue TJ, Dong XZ, Ehrlich DS, Fraser C, Gibbs R, Gilbert J, Gilna P, Glöckner FO, Jansson JK, Keasling JD, Knight R, Labeda D, Lapidus A, Lee JS, Li WJ, Ma J, Markowitz V, Moore ER, Morrison M, Meyer F, Nelson KE, Ohkuma M, Ouzounis CA, Pace N, Parkhill J, Qin N, Rossello-Mora R, Sikorski J, Smith D, Sogin M, Stevens R, Stingl U, Suzuki K, Taylor D, Tiedje JM, Tindall B, Wagner M, Weinstock G, Weissenbach J, White O, Wang J, Zhang L, Zhou YG, Field D, Whitman WB, Garrity GM, Klenk HP (2014) Genomic encyclopedia of bacteria and archaea: sequencing a myriad of type strains. PLoS Biol 12:e1001920
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–175
Liu W-T, Marsh TL, Cheng H, Forney LJ (1997) Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol 63:4516–4522
Liu H, He H, Cheng C, Liu J, Shu M, Jiao Y, Tao F, Zhong W (2014a) Diversity analysis of the bacterial community in tobacco waste extract during reconstituted tobacco process. Appl Microbiol Biotechnol. doi:10.1007/s00253-014-5960-8
Liu J, Hua Z-S, Chen L-X, Kuang J-L, Li S-J, Shu W-S, Huang L-N (2014b) Correlating microbial diversity patterns with geochemistry in an extreme and heterogeneous environment of mine tailings. Appl Environ Microbiol 80:3677–3686
Loman NJ, Constantinidou C, Christner M, Rohde H, Chan JZ, Quick J, Weir JC, Quince C, Smith GP, Betley JR, Aepfelbacher M, Pallen MJ (2013) A culture-independent sequence-based metagenomics approach to the investigation of an outbreak of shiga-toxigenic Escherichia coli O104:H4. JAMA 309:1502–1510
Lu X-M, Lu P-Z (2014) Diversity, abundance, and spatial distribution of riverine microbial communities response to effluents from swine farm versus farmhouse restaurant. Appl Microbiol Biotechnol 98:7597–7608
Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, Von Bergen M, McCoy KD, Macpherson AJ, Danska JS (2013) Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 1(339):1084–1088
Maynaud G, Brunel B, Mornico D, Durot M, Severac D, Dubois E, Navarro E, Cleyet-Marel J-C, Quéré AL (2013) Genome-wide transcriptional responses of two metal-tolerant symbiotic Mesorhizobium isolates to zinc and cadmium exposure. BMC Genomics 14:292
Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilkie A, Wilkening J, Edwards RA (2008) The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinforma 9:386–394
Mohapatra BR, Gould WD, Dinardo O, Koren DW (2011) Tracking the prokaryotic diversity in acid mine drainage-contaminated environments: a review of molecular methods. Miner Eng 24:709–718
Muyzer G, De Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Nies DH (2004) Metals and their compounds in the environment. Part II. In: Anke K, Ihnat M, Stoeppler M (eds) The elements: essential and toxic effects on microorganisms. Wiley-VCH, Weinheim, Germany
Oelofse SHH (2009) Mine water pollution—acid mine decant, effluent and treatment: a consideration of key emerging issues that may impact the state of the environment. In: Krishna CS (ed) Mining: environment and health concerns, 1st edn. The Icfai University Press, India, pp 84–91
Portune KJ, Pérez MC, Álvarez-Hornos FJ, Gabaldón C (2014) Investigating bacterial populations in styrene-degrading biofilters by 16S rDNA tag pyrosequencing. Appl Microbiol Biotechnol. doi:10.1007/s00253-014-5868-3
Quince C, Lanzén A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT (2009) Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods 6:639–641
Raji AI, Moller C, Litthauer D, van Heerden E, Piater LA (2008) Bacterial diversity of biofilm samples from deep mines in South Africa. Biokemistri 20:53–62
Ren G, Zhang H, Lin X, Zhu J, Jia Z (2014) Response of phyllosphere bacterial communities to elevated CO2 during rice growing season. Appl Microbiol Biotechnol 98:9459–9471
Reuter S, Ellington MJ, Cartwright EP, Köser CU, Török ME, Gouliouris T, Harris SR, Brown NM, Holden MT, Quail M, Parkhill J, Smith GP, Bentley SD, Peacock SJ (2013) Rapid bacterial whole-genome sequencing to enhance diagnostic and public health microbiology. JAMA Intern Med 173:1397–1404
Röske I, Sabra W, Nacke H, Daniel R, Zeng A-P, Antranikian G, Sahm K (2014) Microbial community composition and dynamics in high-temperature biogas reactors using industrial bioethanol waste as substrate. Appl Microbiol Biotechnol 98:9095–9106
Shi Y, Yang H, Zhang T, Sun J, Lou K (2014) Illumina-based analysis of endophytic bacterial diversity and space-time dynamics in sugar beet on the north slope of Tianshan mountain. Appl Microbiol Biotechnol 98:6375–6385
Singh B, Crippen TL, Zheng L, Fields AT, Yu Z, Ma Q, Wood TK, Dowd SE, Flores M, Tomberlin JK, Tarone AT (2014) A metagenomic assessment of the bacteria associated with Lucilia sericata and Lucilia cuprina (Diptera: Calliphoridae). Appl Microbiol Biotechnol. doi:10.1007/s00253-014-6115-7
Slonczewski JL, Fujisawa M, Dopson M, Krulwich TA, Robert KP (2009) Cytoplasmic pH measurement and homeostasis in bacteria and archaea. Adv Microb Physiol 55:1–79
Staley C, Unno T, Gould TJ, Jarvis B, Phillips J, Cotner JB, Sadowsky MJ (2013) Application of Illumina next-generation sequencing to characterize the bacterial community of the Upper Mississippi River. J Appl Microbiol 115:1147–1158
Stroud JL, Low A, Collins RN, Manefield M (2014) Metal(loid) bioaccessibility dictates microbial community composition in acid sulfate soil horizons and sulfidic drain sediments. Environ Sci Technol 48:8514–8521
Su X, Zhang Q, Hu J, Hashmi MZ, Ding L, Shen C (2014) Enhanced degradation of biphenyl from PCB-contaminated sediments: the impact of extracellular organic matter from Micrococcus luteus. Microbiol Biotechnol. doi:10.1007/s00253-014-6108-6
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Wang L-Y, Ke W-J, Sun X-B, Liu J-F, Gu J-D, Mu B-Z (2014) Comparison of bacterial community in aqueous and oil phases of water-flooded petroleum reservoirs using pyrosequencing and clone library approaches. Appl Microbiol Biotechnol 98:4209–4221
Whitman WB, Coleman DC, Wiebe WJ (1998) Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A 95:6578–6583
Wu H, Zhang J, Mi Z, Xie S, Chen C, Zhang X (2014) Biofilm bacterial communities in urban drinking water distribution systems transporting waters with different purification strategies. Appl Microbiol Biotechnol. doi:10.1007/s00253-014-6095-7
Ye L, Zhang T (2013) Bacterial communities in different sections of a municipal wastewater treatment plant revealed by 16S rDNA 454 pyrosequencing. Appl Microbiol Biotechnol 97:2681–2690
Zhang Y, Wang X, Hu M, Li P (2014) Effect of hydraulic retention time (HRT) on the biodegradation of trichloroethylene wastewater and anaerobic bacterial community in the UASB reactor. Appl Microbiol Biotechnol. doi:10.1007/s00253-014-6096-6
Zhao D, Huang R, Zeng J, Yu Z, Liu P, Cheng S, Wu QL (2014) Pyrosequencing analysis of bacterial community and assembly in activated sludge samples from different geographic regions in China. Appl Microbiol Biotechnol 98:9119–9128
Acknowledgments
The authors are grateful to the two South African mining companies for allowing the researchers to sample their evaporation dams. The financial support of National Research Foundation (NRF) and the Department of Science and Technology, South Africa for carrying out this project is thankfully acknowledged. JK gratefully acknowledges the Postdoctoral Fellowship from Tshwane University of Technology, Pretoria, South Africa.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 13.8 kb)
Rights and permissions
About this article
Cite this article
Keshri, J., Mankazana, B.B.J. & Momba, M.N.B. Profile of bacterial communities in South African mine-water samples using Illumina next-generation sequencing platform. Appl Microbiol Biotechnol 99, 3233–3242 (2015). https://doi.org/10.1007/s00253-014-6213-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-6213-6