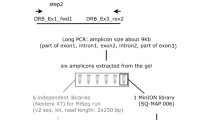
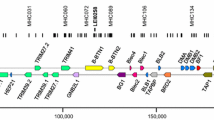
Abstract
Variation in the major histocompatibility complex (MHC) is increasingly associated with disease susceptibility and resistance in avian species of agricultural importance. This variation includes sequence polymorphisms but also structural differences (gene rearrangement) and copy number variation (CNV). The MHC has now been described for multiple galliform species including the best defined assemblies of the chicken (Gallus gallus) and domestic turkey (Meleagris gallopavo). Using this sequence resource, this study applied high-throughput sequencing to investigate MHC variation in turkeys of North America (NA turkeys). An MHC-specific SureSelect (Agilent) capture array was developed, and libraries were created for 14 turkeys representing domestic (commercial bred), heritage breed, and wild turkeys. In addition, a representative of the Ocellated turkey (M. ocellata) and chicken (G. gallus) was included to test cross-species applicability of the capture array allowing for identification of new species-specific polymorphisms. Libraries were hybridized to ∼12 K cRNA baits and the resulting pools were sequenced. On average, 98 % of processed reads mapped to the turkey whole genome sequence and 53 % to the MHC target. In addition to the MHC, capture hybridization recovered sequences corresponding to other MHC regions. Sequence alignment and de novo assembly indicated the presence of several additional BG genes in the turkey with evidence for CNV. Variant detection identified an average of 2245 polymorphisms per individual for the NA turkeys, 3012 for the Ocellated turkey, and 462 variants in the chicken (RJF-256). This study provides an extensive sequence resource for examining MHC variation and its relation to health of this agriculturally important group of birds.



Similar content being viewed by others
References
Abernathy J, Li X, Jia X, Chou W, Lamont SJ, Crooijmans R, Zhou H (2014) Copy number variation in Fayoumi and leghorn chickens analyzed using array comparative genomic hybridization. Anim Genet 45:400–411
Asan XY, Jiang H, Tyler-Smith C, Xue Y, Jiang T, Wang J, Wu M, Liu X, Tian G, Wang J, Wang J, Yang H, Zhang X (2011) Comprehensive comparison of three commercial human whole-exome capture platforms. Genome Biol 12:R95
Aslam ML, Bastiaansen JW, Elferink MG, Megens HJ, Crooijmans RP, le Blomberg A, Fleischer RC, Van Tassell CP, Sonstegard TS, Schroeder SG, Groenen MA, Long JA (2012) Whole genome SNP discovery and analysis of genetic diversity in turkey (Meleagris gallopavo). BMC Genomics 13:391
Bauer MM, Reed KM (2011) Extended sequence of the turkey MHC B-locus sequence variation in the highly polymorphic B-G loci. Immunogenetics 63:209–221
Bauer MM, Miller MM, Briles WE, Reed KM (2013) Genetic variation at the MHC in a population of introduced wild turkeys. Anim Biotechnol 24:210–228
Briles WE, Briles RW (1982) Identification of haplotypes of the chicken major histocompatibility complex (B). Immunogenetics 15:449–459
Briles WE, Bumstead N, Ewert DL, Gilmour DG, Gogusev J, Hala K, Koch C, Longenecker BM, Nordskog AW, Pink JRL, Schierman LW, Simonsen M, Toivanen A, Toivanen P, Vainio O, Wick G (1982) Nomenclature for chicken major histocompatibility (B) complex. Immunogenetics 15:441–447
Butter C, Staines K, van Hateren A, Davison TF, Kaufman J (2013) The peptide motif of the single dominantly expressed class I molecule of the chicken MHC can explain the response to a molecular defined vaccine of infectious bursal disease virus (IBDV). Immunogenetics 65:609–618
Chaves LD, Harry DE, Reed KM (2009a) Genome-wide genetic diversity of "Nici", the DNA source for the CHORI-260 turkey BAC library and candidate for whole genome sequencing. Anim Genet 40:348–352
Chaves LD, Kreuth SB, Reed KM (2009b) Defining the turkey MHC: sequence and genes of the B-locus. J Immunol 183:6530–6537
Chaves LD, Faile GM, Kreuth SB, Hendrickson JA, Reed KM (2010) Haplotype variation, recombination, and gene conversion within the MHC-B of the turkey. Immunogenetics 62:465–477
Chaves LD, Faile GM, Hendrickson JA, Mock K, Reed KM (2011a) A locus-wide approach to assessing variation in the avian MHC: the B-locus of the wild turkey. Heredity 107:40–49
Chaves LD, Kreuth SB, Bauer MM, Reed KM (2011b) Sequence of a turkey BAC clone identifies MHC class III orthologs and supports ancient origins of immunological gene clusters. Cytogenet Genome Res 132:55–63
Chen R, Im H, Snyder M (2015) Whole-exome enrichment with the Agilent SureSelect human all exon platform. Cold Spring Harbor Protoc 2015(7):pdb.prot083659.
Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, Macarthur DG, Macdonald JR, Onyiah I, Pang AW, Robson S, Stirrups K, Valsesia A, Walter K, Wei J, Tyler-Smith C, Carter NP, Lee C, Scherer SW, Hurles ME (2010) Origins and functional impact of copy number variation in the human genome. Nature 464:704–712
Crone M, Simonsen M, Skjodt K, Linnet K, Olsson L (1985) Mouse monoclonal antibodies to class I and class II antigens of the chicken MHC. Immunogenetics 21:181–187
Crooijmans RPMA, Fife MS, Fitzgerald TW, Strickland S, Cheng HH, Kaiser P, Redon R, Groenen MAM (2013) Large scale variation in DNA copy number in chicken breeds. BMC Genomics 14:398. doi:10.1186/1471-2164-14-398
Dalloul RA, Long JA, Zimin AV, Aslam L, Beal K, Blomberg LA, Bouffard P, Burt DW, Crasta O, Crooijmans RP, Cooper K, Coulombe RA, De S, Delany ME, Dodgson JB, Dong JJ, Evans C, Frederickson KM, Flicek P, Florea L, Folkerts O, Groenen MA, Harkins TT, Herrero J, Hoffmann S, Megens HJ, Jiang A, de Jong P, Kaiser P, Kim H, Kim KW, Kim S, Langenberger D, Lee MK, Lee T, Mane S, Marcais G, Marz M, McElroy AP, Modise T, Nefedov M, Notredame C, Paton IR, Payne WS, Pertea G, Prickett D, Puiu D, Qioa D, Raineri E, Ruffier M, Salzberg SL, Schatz MC, Scheuring C, Schmidt CJ, Schroeder S, Searle SM, Smith EJ, Smith J, Sonstegard TS, Stadler PF, Tafer H, Tu ZJ, Van Tassell CP, Vilella AJ, Williams KP, Yorke JA, Zhang L, Zhang HB, Zhang X, Zhang Y, Reed KM (2010) Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biol 8(9). pii: e1000475.
Eimes JA, Reed KM, Mendoza KM, Bollmer JL, Whittingham LA, Bateson ZW, Dunn PO (2013) Greater prairie-chickens have a compact MHC-B with a single class IA locus. Immunogenetics 65:133–144
Fulton JE, Hunt HD, Bacon LD (2001) Chicken major histocompatibility complex class I definition using antisera induced by cloned class I sequences. Poultry Sci 80:1554–1561
Geniez S, Foster JM, Kumar S, Moumen B, Leproust E, Hardy O, Guadalupe M, Thomas SJ, Boone B, Hendrickson C, Bouchon D, Grève P, Slatko BE (2012) Targeted genome enrichment for efficient purification of endosymbiont DNA from host DNA. Symbiosis 58:201–207
Giuffre A, Pabon-Pena C, Novak B, Joshi S, Ong J, Visitacion M, Hamady M, Useche F, Eberle J, Hunt S, Happe S, Roberts D, Leproust E (2011) The Agilent Technologies’ SureSelectTM All Exon Product Portfolio: High performance target enrichment system for human and mouse exome sequencing on Illumina and SOLiD platforms. J Biomol Tech 22(Suppl):S41
Gonzalez-Quevedo C, Phillips KP, Spurgin LG, Richardson DS (2015) 454 screening of individual MHC variation in an endemic island passerine. Immunogenetics 67:149–162
Goto RM, Wang Y, Taylor RL, Wakenell PS, Hosomichi K, Shiina T, Blackmore CS, Briles WE, Miller MM (2009) BG1 has a major role in MHC-linked resistance to malignant lymphoma in the chicken. Proc Natl Acad Sci U S A 106:16740–16745
Griffin DK, Robertson LB, Tempest HG, Vignal A, Fillon V, Crooijmans RP, Groenen MA, Deryusheva S, Gaginskaya E, Carre W, Waddington D, Talbot R, Volker M, Masabanda JS, Burt DW (2008) Whole genome comparative studies between chicken and turkey and their implications for avian genome evolution. BMC Genomics 9:168
Groenen MAM, Megens H-K, Zare Y, Warren WC, Hillier LW, Crooijmans RPMA, Vereijken A, Okimoto R, Muir WM, Cheng HH (2011) The development and characterization of a 60K SNP chip for chicken. BMC Genomics 12:274
Guo Y, Long J, He J, Li CI, Cai Q, Shu XO, Zheng W, Li C (2012) Exome sequencing generates high quality data in non-target regions. BMC Genomics 13:194
Han R, Yang P, Tian Y, Wang D, Zhang Z, Wang L, Li Z, Jiang R, Kang X (2014) Identification and functional characterization of copy number variations in diverse chicken breeds. BMC Genomics 15:934
Hofmann A, Plachy J, Hunt L, Kaufman J, Hala K (2003) v-src oncogene-specific carboxy-terminal peptide is immunoprotective against Rous sarcoma growth in chickens with MHC class I allele B-F12. Vaccine 21:4694–4699
Hosomichi K, Miller MM, Goto RM, Wang Y, Suzuki S, Kulski JK, Nishibori M, Inoko H, Hanzawa K, Shiina T (2008) Contribution of mutation, recombination, and gene conversion to chicken MHC-B haplotype diversity. J Immunol 181:3393–3399
Hunt HD, Dunn JR (2013) The influence of host genetics on Marek’s disease virus evolution. Avian Dis 57:474–482
Jia X, Chen S, Zhou H, Li D, Liu W, Yang N (2013) Copy number variations identified in the chicken using a 60 K SNP BeadChip. Anim Genet 44:276–284
Kaufman J (2008) The avian MHC. In: Davison TF, Kaspers B, Schat KA (eds) Avian immunology. Elsevier, Ltd, London, pp 159–182
Kaufman J, Salomonsen J, Skjødt K (1989) B-G cDNA clones have multiple small repeats and hybridize to both chicken MHC regions. Immunogenetics 30:440–451
Kaufman J, Milne S, Göbel TW, Walker BA, Jacob JP, Auffray C, Zoorob R, Beck S (1999) The chicken B locus is a minimal essential major histocompatibility complex. Nature 401:923–925
Kranis A, Gheyas AA, Boschiero C, Turner F, Yu L, Smith S, Talbot R, Pirani A, Brew F, Kaiser P, Hocking PM, Fife M, Salmon N, Fulton J, Strom TM, Haberer G, Weigend S, Preisinger R, Gholami M, Qanbari S, Simianer H, Watson KA, Woolliams JA, Burt DW (2013) Development of a high density 600K SNP genotyping array for chicken. BMC Genomics 14:59
Liao GJ, Lun FM, Zheng YW, Chan KC, Leung TY, Lau TK, Chiu RW, Lo YM (2011) Targeted massively parallel sequencing of maternal plasma DNA permits efficient and unbiased detection of fetal alleles. Clin Chem 57:92–101
Longenecker BM, Mosmann TR (1980) Restricted expression of an MHC alloantigen in cells of the erythroid series: a specific marker for erythroid differentiation. J Supramol Struct 13:395–400
Luo J, Yu Y, Mitra A, Chang S, Zhang H, Liu G, Yang N, Song J (2013) Genome-wide copy number variant analysis in inbred chickens lines with different susceptibility to Marek’s disease. G3 (Bethesda) 3:217–223.
McCarroll SA, Altshuler DM (2007) Copy-number variation and association studies of human disease. Nat Genet 39:S37–42
Monson MS, Mendoza KM, Velleman SG, Strasburg GM, Reed KM (2013) Expression profiles for genes in the avian MHC B-Locus. Poult Sci 92:1523–1534
Pink JRL, Kieran WM, Rijnbeek M-A, Longenecker BM (1985) A monoclonal antibody against chicken MHC class I (B-F) antigens. Immunogenetics 21:293–297
Reed KM, Bauer MM, Monson MS, Chaves LD, Benoit B, O’Hare TH, Delany ME (2011) Defining the turkey MHC: identification of expressed class I and class II-like genes independent of the B-locus. Immunogenetics 63:753–771
Reed KM, Benoit B, Wang X, Greenshields M, Hughes C, Mendoza KM (2014) Conserved MHC gene orthologs genetically map to the turkey MHC-B. Cytogenet Genome Res 144:31–38
Salomonsen J, Chattaway JA, Chan ACY, Parker A, Huguet S, Marston DA, Rogers SL, Wu Z, Smith AL, Staines K, Butter C, Riegert P, Vainio O, Nielsen L, Kaspers B, Griffin DK, Yang F, Zoorob R, Guillemot F, Auffray C, Beck S, Skjødt K, Kaufman J (2014) Sequence of a complete chicken BG haplotype shows dynamic expansion and contraction of two gene lineages with particular expression patterns. PLoS Genet 10:e1004417
Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, Pertz LM, Clark RA, Schwartz S, Segraves R, Oseroff VV, Albertson DG, Pinkel D, Eichler EE (2005) Segmental duplications and copy-number variation in the human genome. Am J Hum Genet 77:78–88
Shaw I, Powell TJ, Marston DA, Baker K, van Hateren A, Riegert P, Wiles MV, Milne S, Beck S, Kaufman J (2007) Different evolutionary histories of the two classical class I genes BF1 and BF2 illustrate drift and selection within the stable MHC haplotypes of chickens. J Immunol 178:5744–52
Shiina T, Shimizu S, Hosomichi K, Kohara S, Watanabe S, Hanzawa K, Beck S, Kulski JK, Inoko H (2004) Comparative genomic analysis of two avian (quail and chicken) MHC regions. J Immunol 172:6751–6763
Shiina T, Briles WE, Goto RM, Hosomichi K, Yanagiya K, Shimizu S, Inoko H, Miller MM (2007) Extended gene map reveals tripartite motif, C-type lectin, and Ig superfamily type genes within a subregion of the chicken MHC-B affecting infectious disease. J Immunol 78:7162–7172
Spurgin LG, van Oosterhout C, Illera JC, Bridgett S, Gharbi K, Emerson BC, Richardson DS (2011) Gene conversion rapidly generates major histocompatibility complex diversity in recently founded bird populations. Mol Ecol 20:5213–5225
Tian M, Wang Y, Gu X, Feng C, Fang S, Hu X, Li N (2013) Copy number variants in locally raised Chinese chicken genomes determined using array comparative genomic hybridization. BMC Genomics 14:262
Usher CL, McCarroll SA (2015) Complex and multi-allelic copy number variation in human disease. Brief Funct Genomics 14:329–338
Wang X, Nahashon S, Feaster TK, Bohannon-Stewart A, Adefope N (2010) An initial map of chromosomal segmental copy number variations in the chicken. BMC Genomics 11:351
Wang B, Ekblom R, Strand TM, Portela-Bens S, Höglund J (2012a) Sequencing of the core MHC region of black grouse (Tetrao tetrix) and comparative genomics of the galliform MHC. BMC Genomics 13:553
Wang Y, Gu X, Feng C, Song C, Hu X, Li N (2012b) A genome-wide survey of copy number variation regions in various chicken breeds by array comparative genomic hybridization method. Anim Genet 43:282–289
Yan Y, Yi G, Sun C, Qu L, Yang N (2014) Genome-wide characterization of insertion and deletion variation in chicken using next generation sequencing. PLoS One 9(8):e104652
Yan Y, Yang N, Cheng HH, Song J, Qu L (2015) Genome-wide identification of copy number variations between two chicken lines that differ in genetic resistance to Marek’s disease. BMC Genomics 16:843
Yi G, Qu L, Liu J, Yan Y, Xu G, Yang N (2014) Genome-wide patterns of copy number variation in the diversified chicken genomes using next-generation sequencing. BMC Genomics 15:962
Yi G, Qu L, Chen S, Xu G, Yang N (2015) Genome-wide copy number profiling using high-density SNP array in chickens. Anim Genet 46:148–157
Zhou W, Liu R, Zhang J, Zheng M, Li P, Chang G, Wen J, Zhao G (2014) A genome-wide detection of copy number variation using SNP genotyping arrays in Beijing-You chickens. Genetica 142:441–450
Acknowledgments
The authors thank Kevin Willis, Minnesota Zoo and Chris Brown, Dallas Zoo for assistance in obtaining blood samples of Ocellated turkey; Ed Smith (VT) and Karen Mock (USU) for supplying Heritage and wild turkey DNA; and Jerry Dodgson (MSU) for supplying chicken DNA (RJF-256). This study was funded in part by the University of Minnesota Agricultural Experiment Station. The authors acknowledge the Minnesota Supercomputing Institute (MSI) at the University of Minnesota for providing resources that contributed to the research results reported within this paper.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
Consensus DNA sequences for mapped reads extracted from four MHC-B alignment peaks for Nici. (DOCX 42 kb)
ESM 2
Aligned nucleotide and amino acid sequence corresponding to BG exon 2 variants observed in contigs assembled from sequence reads for Nici mapped to the B12 BG region (KC955130). (DOCX 22 kb)
ESM 3
Table 1 Summary of BLAST results for contigs assembled from Nici sequence reads that did not map to the turkey genome sequence. Included for each contig are the size (bp), lowest observed E value, accession number, and description of the best match. (XLSX 21 kb)
ESM 4
Table 2 MHC sequence variants observed in the control bird Nici. For each variant, the position, type, length, reference sequence, observed allele, zygosity, count, coverage, frequency, average quality, and flanking sequence are given. Positions are given for the compiled MHC-B reference (Genbank nos. HQ008883 and DQ993255). Variant types are as follows: SNV = single nucleotide, MNV = multiple nucleotide and Indel = insertion deletions. (XLSX 15 kb)
ESM 5
Table 3 MHC sequence variants for turkey (Meleagris gallopavo). For each variant, the reference position, position in the corresponding BAC clone, variant type, reference sequence, observed allele, zygosity, count, coverage, frequency, average quality, and gene are given. Positions are listed for the compiled MHC-B (Genbank nos. HQ008883 and DQ993255 reference) and individual BAC clones assemblies. Variant types are as follows: SNV = single nucleotide, MNV = multiple nucleotide, and Indel = insertion deletions. Red text indicates SNPs verified from previous studies (Chaves et al. 2010; 2011b). (XLSX 628 kb)
ESM 6
Table 4 Sequence variants observed in the Ocellated turkey (Meleagris ocellata). For each variant, the position, variant type, reference sequence, observed variant(s), and allele frequencies are given. Positions are relative to the MHC-B of M. gallopavo as denoted in Supplementary Table 2. Variant types are as follows: SNV = single nucleotide, MNV = multiple nucleotide, and Indel = insertion deletions. (XLSX 226 kb)
ESM 7
Table 5 Sequence variants observed in the chicken (RJF-256). For each variant, the position, variant type, reference sequence, observed variant(s), and allele frequency are given. Positions are as found for the MHC assembly (Genbank no. AB268588). Variant types are as follows: SNV = single nucleotide, MNV = multiple nucleotide, and Indel = insertion deletions. (XLSX 51 kb)
Rights and permissions
About this article
Cite this article
Reed, K.M., Mendoza, K.M. & Settlage, R.E. Targeted capture enrichment and sequencing identifies extensive nucleotide variation in the turkey MHC-B . Immunogenetics 68, 219–229 (2016). https://doi.org/10.1007/s00251-015-0893-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-015-0893-7