Abstract
Proteolysis is the general term to describe the process of protein degradation into peptides. Proteasomes are the main actors in cellular proteolysis, and their activity can be measured in in vitro digestion experiments. However, in vivo proteolysis can be different than what is measured in these experiments if other proteases participate or if proteasomal activity is different in vivo. The in vivo proteolysis can be measured only indirectly, by the analysis of peptides presented on MHC-I molecules. MHC-I presented peptides are protected from further degradation, thus enabling an indirect view on the underlying in vivo proteolysis. The ligands presented on different MHC-I molecules enable different views on this process; in combination, they might give a complete picture. Based on in vitro proteasome-only digestions and MHC-I ligand data, different proteolysis predictors have been developed. With new in vitro digestion and MHC-I ligand data sets, we benchmarked how well these predictors capture in vitro proteasome-only activity and in vivo whole-cell proteolysis, respectively. Even though the in vitro proteasome digestion patterns were best captured by methods trained on such data (ProteaSMM and NetChop 20S), the in vivo whole-cell proteolysis was best predicted by a method trained on MHC-I ligand data (NetChop Cterm). Follow-up analysis showed that the likely source of this difference is the activity from proteases other than the proteasome, such as TPPII. This non-proteasomal in vivo activity is captured by NetChop Cterm and should be taken into account in MHC-I ligand predictions.
Similar content being viewed by others
Introduction
The proteasome degrades intracellular proteins, marked for degradation by the ubiquitination pathway (Hershko and Ciechanover 1992). Protein degradation, i.e., proteolysis, is important to remove miss-folded proteins, to regulate cellular processes such as the cell-cycle or for the production of MHC-I ligands (Goldberg 2003; Seifert et al. 2010; van Leuken et al. 2008; Clijsters et al. 2013; Kloetzel 2001). Peptide fragments that result from proteolysis are rapidly degraded by cytosolic aminopeptidases (Reits et al. 2003). However, few peptides escape this degradation and are transported to the endoplasmatic reticulum (ER) by the transporter associated with antigen processing (TAP), where they can form peptide-MHC-I complexes (pMHCs) (Neefjes et al. 2011). pMHCs are presented on the cell surface to enable immune surveillance by T cells.
Most cells express the constitutive proteasome, which is a barrel-shaped multi-subunit protein complex, composed of two α- and two β-rings, where each ring contains seven subunits. In the β ring of the constitutive proteasome, three proteins are present that have proteolytic capacity: β1, β2, and β5 (Kloetzel and Ossendorp 2004). Under the influence of interferon- γ (IFN γ), these subunits can be substituted by β1 i , β2 i , and β5 i , respectively, to form the so-called immunoproteasome (Aki et al. 1994). Whereas the constitutive proteasome has a preference to cleave hydrophobic, acidic, and basic amino acids, the immunoproteasome is more efficiently cleaving after hydrophobic and basic amino acids (Gaczynska et al. 1993; Toes et al. 2001; Kesmir et al. 2003). Other proteasome types can be formed by a combination of constitutive and immunoproteasomal subunits (Guillaume et al. 2010), or with the β5 t subunit that is only expressed in cortical thymic epithelial cells (Murata et al. 2007). These different proteasome types largely overlap in their cleavage preferences (Guillaume et al. 2010; Murata et al. 2007; Florea et al. 2010), though the efficiency can differ at different cleavage sites which has an influence on the repertoire of MHC-I presented peptides (Kincaid et al. 2012).
Two main approaches have been taken to study proteolytic activity: in vitro digestion experiments and in vivo MHC-I-ligand elutions. In an in vitro digestion experiment, a protein is incubated with proteasomes. The peptide fragments that are formed during the digestion can be detected by mass spectrometry, and cleavage sites can be inferred from the fragments (Emmerich et al. 2000; Tenzer et al. 2004; Toes et al. 2001). So far, the cleavage sites of only three proteins, i.e., β-casein, enolase, and prion protein, have been determined in such in vitro assays (Emmerich et al. 2000; Tenzer et al. 2004; Toes et al. 2001). Alternatively, in vivo proteolytic activity can be measured by the analysis of digestion fragments that form pMHCs; these fragments can be eluted from a cell and identified by mass spectrometry. The C-terminus of the MHC-I presented peptide is generated by proteolytic activity and reflects an in vivo cleavage site in the protein from which the MHC-I ligand was derived (Kloetzel 2001). However, as many cleavage sites will result in fragments that do not become MHC-I ligands, only a small subset of all cleavage sites can be detected via this approach. In addition, other peptidases such as ACE, TPPII, and Nardilysin (Geier et al. 1999; Shen et al. 2008; Kessler et al. 2011) can influence the C-terminus of MHC-I ligands. Therefore, the MHC-I-ligand data is more likely to reflect the proteolytic activity of all cellular proteases, rather than the activity of just the proteasomes or one proteasome-type.
To study proteolyis and to aid MHC-I ligand predictions, different proteolyis predictors have been developed (Holzhutter et al. 1999; Holzhutter and Kloetzel 2000; Kesmir et al. 2002; Nielsen et al. 2005; Tenzer et al. 2005; Ginodi et al. 2008; Kuttler et al. 2000; Nussbaum et al. 2001. Most predictors, e.g., FragPredict (Holzhutter et al. 1999; Holzhutter and Kloetzel 2000), ProteaSMM (Tenzer et al. 2005), PAProC (Kuttler et al. 2000; Nussbaum et al. 2001), and PepCleave (Ginodi et al. 2008), have been trained on the in vitro proteasome digestion data from β-casein and enolase (Emmerich et al. 2000; Toes et al. 2001). NetChop 20S (Kesmir et al. 2002; Nielsen et al. 2005) and the so-called enhanced versions of ProteaSMM are trained on the in vitro proteasome digestion data from β-casein, enolase, and the prion-protein (Emmerich et al. 2000; Toes et al. 2001; Tenzer et al. 2004). Unlike the other predictors, NetChop Cterm is trained on in vivo MHC-I ligand data (Kesmir et al. 2002; Nielsen et al. 2005). Besides the different data sets that were used for training the methods, different computational techniques were used to construct the predictors. For instance, ProteaSMM models the cleavage pattern with a stabilized matrix method (SMM) using six amino acids C-terminal and four amino acids N-terminal of a potential cleavage site, and NetChop is based on a neural network that uses nine amino acids C-terminal and eight amino acids N-terminal of a potential cleavage site.
In 2005, Tenzer et al. (2005) bench-marked FragPredict, PAProC, NetChop-2.0, and ProteaSMM on several data sets, and showed that ProteaSMM best predicted in vitro proteasome digestion cleavage patterns, whereas NetChop-2.0 Cterm best predicted the cleavage patterns based on MHC-I ligands. Tenzer et al. (2005) argued that the increased performance of NetChop-2.0 Cterm on the MHC-I ligand data was due to a recognition of TAP-transportable peptides. After this study, NetChop was updated to version 3.0 (Nielsen et al. 2005) and a new method, PepCleave, was developed (Ginodi et al. 2008). Unfortunately, PepCleave cannot be compared to the other predictors as it predicts fragments and not cleavages (Ginodi et al. 2008). Therefore, we have chosen to compare ProteaSMM and the newest version of NetChop on new in vitro proteasome digestion data sets, and a new benchmark set of MHC-I ligands. Next to benchmarking, our analysis shines light on the nature of the difference between in vitro proteasome-only and in vivo whole-cell proteolytic activities, suggesting an important role for proteases other than the proteasome.
Results
Predicting in vitro cleavage patterns
To compare proteasome predictors, we generated a new independent data set. This data set was based on in vitro digestions of 17–30 amino acids long HIV–1 peptides; the products of these digestions were analyzed using mass spectrometry to determine cleavage and non-cleavage sites (see “Methods” section). Digestions were performed with either constitutive or immunoproteasomes. Of 368 possible cleavage sites, 150 (41 %) were used by the constitutive proteasomes, and 148 by the immunoproteasomes, 103 sites (of the 148 cleavage sites) were cleaved by both proteasome-types (Supplementary Table S1). Thus, even though the different proteasomes can target the different sites with varying efficiencies, the set of cleavage sites that is identified in this assay largely overlaps.
The prediction performance of ProteaSMM and NetChop-3.0 was analyzed using receiver operator characteristic (ROC) curves, where the number of correct and false predictions is plotted for every prediction threshold (Swets 1988). The area under a ROC-curve (AUC) is a performance measure of the predictor, and is widely used because it is threshold independent (Swets 1988). For each predictor (and different versions of the predictors), the AUCs were determined on both constitutive and immunoproteasomal cleavage patterns obtained from the in vitro digestions (Table 1). In general, the methods performed better in predicting the immunoproteasomal cleavage pattern. This could be explained by the more biased cleavage preference of immunoproteasomes, that cleave after hydrophobic and basic amino acids with greater, and after acidic amino acids with lesser efficiency (Gaczynska et al. 1993; Toes et al. 2001; Kesmir et al. 2003). Such a more biased cleavage pattern might be easier to predict. The immunoproteasomal cleavage pattern was best predicted by proteaSMM-immuno and proteaSMM-constitutive (ROC-comparison test: p < 0.001; Table 1), and the constitutive cleavage pattern was best captured by proteaSMM-constitutive and NetChop-3.0 20S (ROC-comparison test: p < 0.001; Table 1). Surprisingly, the enhanced ProteaSMM versions did not perform better, even though they are trained on extra data from proteasomally digested prion protein (Tenzer et al. 2004). NetChop-3.0 20S is also trained on prion data, but no version of this method is available that is not trained on prion data, to test if prion data negatively affects the performance of NetChop-3.0 20S.
In summary, the methods that have been trained on in vitro proteasome digestion data (proteaSMMs and NetChop-3.0 20S) outperformed the method that has been trained on in vivo MHC-I ligand data (NetChop-3.0 Cterm), which agrees with previous observations (Tenzer et al. 2005; Saxova et al. 2003) and the expectation that methods trained on in vitro data can best predict proteasome-only cleavage patterns.
Predicting in vivo cleavage patterns
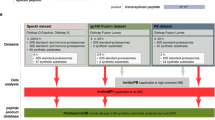
In vivo proteolytic activity can be rather different from pure proteasomal activity, if other peptidases e.g., ACE, TPPII, or Nardilysin (Geier et al. 1999; Shen et al. 2008, 2011; Kessler et al. 2011) contribute to the in vivo proteolysis. As a result, the ability of different proteasome predictors to predict in vivo proteolysis might be different from their ability to predict in vitro proteasome-only cleavages. To test and compare the in vivo proteolysis prediction performances, we inferred in vivo cleavage sites from non-redundant MHC-I ligands that have been identified from 2005 on, after NetChop Cterm was last updated (n = 832; see Fig. 1 and “Methods” section). A data set of in vivo non-cleavage sites was derived in two ways: (1) by shuffling, 100 non-cleavage sites were made by shuffling the 19 amino acids flanking a cleavage site (the area used by NetChop for predictions plus one N-terminal and one C-terminal extension, see Fig. 2). (2) By assuming that all sites in the source protein of the MHC-I ligand that are not identified as cleavage sites are non-cleavage sites (“Methods” section). The predictors were assessed for their capacity to discriminate cleavage sites from non-cleavage sites, by comparing AUC values. Not surprisingly, NetChop-3.0 Cterm most accurately captured the in vivo cleavage pattern irrespective of the non-cleavage data set (ROC-comparison test: p < 0.001; Fig. 3). This is expected as NetChop Cterm has been trained on in vivo cleavage patterns inferred from MHC-I ligands.
Constructing the MHC-I ligand data set. MHC-I ligands and source proteins, that were discovered in elution studies, were derived from the SYFPHEITI database (Rammensee et al. 1999) and the IEDB database (Vita et al. 2010). The data sets were combined and non-redundant ligands that were not published before 2005 were selected. Every MHC-I ligand in its source protein represents a cleavage site; non-cleavage sites were derived by either shuffling an area of 19 amino acids around the cleavage site (Fig. 2 and “Methods” section), or by defining all other sites in the source proteins of MHC-I ligands as non-cleavage sites (“Methods” section)
Constructing non-cleavage sites by shuffling. The C-terminus of an MHC-I ligand (between P1’ and P1) is defined as a cleavage site. An area of 19 amino acids (from P10’ to P9) around a cleavage site was shuffled and the middle position was assigned as a non-cleavage site. For every cleavage site, 100 non-cleavage sites were constructed. The positions that are used by NetChop-3.0 (P9’ to P8) and ProteaSMM (P6’ to P4) for predicting cleavage probabilities are indicated
Predicting in vivo proteolysis. Proteasome cleavage predictors were tested as a stand-alone predictor, or in combination with a TAP predictor, and performance was assessed using AUC and VUP, respectively (see “Methods” section). The performance was tested using either non-cleavage data sets derived by the shuffling method (a) or by taking other sites from the source protein as non-cleavage sites (b) (see Fig. 1 and “Methods” section). Examples of the AUC and the VUP analyses are shown in the upper part, AUC and VUP scores are given in the lower part. In all analyses, NetChop-3.0 Cterm showed the highest performance (ROC-comparison test: p <0.001). The performance of NetChop-3.0 Cterm is shown in red lines, NetChop-3.0 20S in black, ProteaSMM Immuno in yellow, ProteaSMM Immuno enhanced in green, ProteaSMM constitutive in blue, and ProteaSMM constitutive enhanced in magenta lines
As in vivo proteolysis is inferred from MHC-I ligand data and NetChop Cterm is trained on such data, Tenzer et al. (2005) noted in an earlier benchmark study that the superior performance of NetChop might be due to a biased recognition of peptides with a high TAP affinity. To exclude this effect, the performance of the different proteasome predictors was tested in combination with a TAP transport predictor (Peters et al. 2003). Therefore, we first followed the approach from Tenzer et al. (2005) by summing TAP-transport and proteasome cleavage scores into a single score. For both non-cleavage site definitions, NetChop-3.0 Cterm outperformed the other predictors, even when the TAP transport scores were differently weighted prior to summation (Fig. 4 and S1).
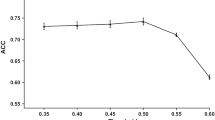
Predicting in vivo proteolysis by combining proteolysis and TAP transport predictor scores. For the different proteasome cleavage predictors, the proteasome cleavage prediction score was added to the TAP transport prediction score (as proposed by Tenzer et al. (2005)). Prediction performance was measured as the AUC of an ROC-curve (Y-axis), using the shuffled sequences as non-cleavage sites (see Fig. 2 and “Methods” section). When combining the scores, the weight of the TAP transport score was changed by the factor W (on the X-axis). The combined score (C), based on the TAP transport (T) and proteolysis (P) score would be C=W∗T+P. As a result, the proteasome cleavage or the TAP transport predictor has a larger influence in the combined score if W is smaller or larger, respectively. See Fig. 3 for color coding
In an AUC-analysis, one can test the predictive performance of a single set of scores. However, we wanted to test the performance of a combination of two scores, i.e., proteasome cleavage and TAP transport scores, as an alternative to the additive model proposed by as Tenzer et al. did. Therefore, we developed a new method to measure the performance of these two scores simultaneously. In this method, for every TAP binding threshold, the performance of the cleavage predictor was measured on cleavage and non-cleavage sites exceeding the threshold. Next, an integration over all the performance scores was combined in a score called volume under the plane (VUP; see “Methods” section). For both non-cleavage definitions, NetChop-3.0 Cterm outperformed the other proteasome predictors based on VUP-scores (ROC-comparison test: p < 0.001; Fig. 3), again indicating that its higher performance is not due to a biased recognition of TAP ligands. Taken together, NetChop Cterm seems to predict in vivo proteolysis better than the other predictors that are trained on proteasome-only in vitro proteolysis data. This suggests that the proteolytic activity in vivo that underlies MHC-I ligand production is markedly different from in vitro proteasome-only proteolysis.
Comparing in vitro and in vivo proteolysis activity
To better understand why NetChop-3.0 Cterm predicts in vivo proteolysis better than the other predictors, even though these predictors better predict proteasome-only in vitro proteolysis, we examined for each predictor which cleavage sites were given a low prediction score. The cleavage sites with a bottom 5 % prediction score were selected for further analysis. A striking difference between NetChop-3.0 Cterm and the other predictors was observed at position P1’ of these poorly predicted cleavage sites (i.e., the C-terminus of the MHC-I ligand; Fig. 2). Whereas the amino acids at position P1’ were equally distributed for NetChop-3.0 Cterm, a Lysine was found in at least 50 % of the cases for the other predictors (Fig. 5). In other words, the predictors based on in vitro proteasomal cleavage data fail to capture the in vivo cleavage after Lysine residues. This fits with the described proteolytic preferences of TPPII and Nardilysin (Geier et al. 1999; Kessler et al. 2011), and the suggested role of these proteases in the generation of MHC-I ligands, for instance for HLA-A*03 and HLA-A*11 (Seifert et al. 2003; Kloetzel 2004; Kloetzel and Ossendorp 2004. In addition, other proteases such as ACE have been shown to influence the generation of MHC-I ligands (Shen et al. 2008, 2011) and their proteolytic activity could be captured by NetChop-3.0 Cterm. Taken together, these results suggest that NetChop-3.0 Cterm incorporates the activity of all different proteases that make a substantial contribution to in vivo proteolysis, thereby can predict in vivo proteolysis better.
Proteolytic activity after Lysine residues is only predicted by NetChop-3.0 Cterm. For every proteasome cleavage predictor, 5 % of the true cleavage sites with the lowest prediction scores were determined. The amino acid profile at P1’ (i.e., the C-terminus of the presented MHC-I ligand) of these cleavage sites with a low prediction score was analyzed. The height of the letters represents their frequency in the amino acid profile
Discussion
In this study, we analyzed how well different methods can predict the cleavage patterns in proteolysis. In vitro cleavage patterns were shown to be best captured by methods trained on in vitro proteasome digestion data, i.e., ProteaSMM and NetChop-3.0 20S (Table 1). Similarly, in vivo proteolysis was best predicted by the method that is trained on MHC-I ligand data, NetChop-3.0 Cterm (Fig. 3). Furthermore, we showed that the better prediction of in vivo proteolysis was not due to an embedded recognition of TAP transportable peptides (Figs. 3 and 4).
There can be two explanations for the difference between in vitro and in vivo proteolysis: First, the proteolytic activity of proteasomes in vitro might be different from their in vivo activity. This difference might result from the interactions with other molecules such as PA28 or the 19S cap regulatory particle (de Graaf et al. 2011; Emmerich et al. 2000). Second, other proteases such as TPPII, ACE, or Nardilysin might make a substantial contribution to the in vivo proteolysis (Geier et al. 1999; Shen et al. 2008, 2011; Kessler et al. 2011). The best described example of in vivo proteolytic activity that is not observed in vitro is the cleavage after Lysine residues. This activity is required to generate ligands for HLA–A*03 and HLA–A*11 that bind peptides with a Lysine at the C-terminus (Seifert et al. 2003; Kloetzel 2004; Kloetzel and Ossendorp 2004). A well-described example of such peptides is the HIV Nef-derived epitope at positions 73 to 82 with a Lysine at its C-terminus, and it was shown that the generation of this peptide depends on TPPII activity (Seifert et al. 2003). However, it is not yet known how dominant this endopeptidase activity is within the TPPII enzyme complex (Geier et al. 1999), and therefore it is not yet clear whether TPPII is responsible for all the activities creating the peptides with a Lysine at its C-terminal. More recently, a more detailed analysis of the substrate specificity of TPPII has been published , which suggests that the endopeptidase activity of TPPII is very much dependent on the length of the substrate and thus is not likely to be a very general enzymatic activity of TPPII. We show that only NetChop-3.0 Cterm captures this hallmark of in vivo proteolysis (Fig. 5). As this activity has not been contributed to the proteasome, we conclude that NetChop Cterm has learned to incorporate non-proteasomal proteolytic activity.
A biased recognition of TAP transportable peptide is not explaining the increased performance of NetChop Cterm on the prediction of in vivo cleavage sites derived from MHC-I ligand data (Figs. 4 and 5). Similarly, one could think that a bias to recognize MHC-I presented ligands should be controlled. NetChop Cterm, it was trained on in vivo cleavage sites derived from a set of pMHCs with a homogenous distribution of MHC-I molecules with various binding preferences (Nielsen et al. 2005) to minimize such a bias that would be due to the recognition of MHC-I binding peptides. In addition, the in vivo cleavage/non-cleavage site data sets in this study are derived from peptides that were not used to train NetChop Cterm and that were eluted from many different MHC-I molecules.
The evaluation of different proteasome cleavage predictors depends on the construction of a set of non-cleavage sites, as the performance on these and on the true cleavage sites needs to be compared. Unfortunately, a substantial set of true non-cleavage sites is not available, and therefore we have to rely on assumptions when compiling a set of non-cleavage sites. To prevent a bias as a result of such assumptions, we have followed two different sets of assumptions when constructing the non-cleavage sites. First, non-cleavage sites were made by shuffling the sequence around a cleavage site to destroy any motif that is used by the proteasome while keeping the same distribution of amino acids. Second, we considered other positions in the source protein as non-cleavage sites. Although identical conclusions were drawn from the analyses with the different sets of non-cleavage sites, identification of true in vivo non-cleavage sites is required to permanently settle this issue or to describe sequence motifs that truly inhibit proteasomal cleavage.
The development of proteasome predictors serves two goals. First, to understand the specificity and biochemical processes that underly proteolysis. Second, to predict and understand how this process influences the MHC-I ligandome. With respect to the first goal, we show that profound differences between proteasome activity in vitro and cellular proteolysis in vivo exist, suggesting a non-negligible role of non-proteasomal proteases. Evidently, the specificity of these additional proteases should be taken into account for optimal MHC-I ligand predictions. Therefore, we conclude that NetChop Cterm or future proteolysis predictors trained on in vivo data should be used in MHC-I ligandome predictions.
Methods
Data collection
Proteasomal in vitro cleavage patterns were derived from a digestion of HIV-1 peptides with constitutive or immuno-proteasomes, as explained in (Peters et al. 2002). Sixteen peptides from the HIV-1 proteins GAG and TAT, with a length of 17 to 30 amino acids, were degraded. After 0, 1, 2, 4, 8, and 24 h of degradation, peptide fragments were analyzed using mass spectrometry (as in Peters et al. (2002)). To avoid analyzing secondary cleavage products, peptide fragments found after 4 h of degradation were used to infer cleavage sites. Of 368 possible cleavage sites, 150 were efficiently cleaved by the immunoproteasome after 4 h and 148 were efficiently cleaved by the constitutive proteasome; 103 sites (69 %) were shown to be cleaved by both proteasome subtypes (Supplementary Table S1). The ProteaSMM proteasome cleavage predictors require six amino acids N-terminal and four amino acids C-terminal of a possible cleavage site. Therefore, cleavage predictions cannot be made at the beginning and end of a peptide sequence. As a result of this limitation, only 240 (of the 368) sites could be used to compare the different proteasome predictions. Of these 240 sites, 99 were efficiently cleaved by the immunoproteasome and 99 were efficiently cleaved by the constitutive proteasome; 68 sites were put in both sets.
In vivo cleavage sites were inferred from MHC-I ligand data . Ligands that were identified in MHC-I elution studies were downloaded from the SYFPHEITI database (Rammensee et al. 1999) and the IEDB database (Vita et al. 2010). Source proteins of the MHC-I ligands were downloaded from the NCBI via links that were provided by the SYFPHEITI and IEDB databases. The C-terminal residue of an MHC-I ligand was regarded as position P1’ of a cleavage site (Fig. 2). In total, 3076 MHC-I ligands with their source protein were derived from the SYFPHEITI database and 457 MHC-I ligands with their source protein were derived from the IEDB database. Identical peptides, or peptides that were either a C- or N-terminal extension of each other, were regarded as redundant. In addition, the ligands and their corresponding source proteins that were published before 2005, or which were redundant/identical to an MHC-I ligand published before 2005, were excluded because they could have been used for training of NetChop-3.0 Cterm. This filtering resulted in 832 MHC-ligands and their source proteins, of which every MHC-I ligand corresponds to a peptide fragment that is generated by in vivo proteolytic activity (Fig. 1).
Detecting in vivo non-cleavage sites based on the absence of a peptide in the MHC-I ligand databases is not possible, as many other reasons might underlie the absence of an MHC-I ligand, e.g., further degradation of the fragment or low affinity to MHC-I molecules. Therefore, non-cleavage sites were generated in two ways: (1) by shuffling of an area of 19 amino acids around the cleavage site (the longest flanking region used by a proteasome predictor method plus one extra amino acid on each side, as indicated in Fig. 2). After shuffling, the middle position, previously corresponding to the cleavage site, was assigned as a non-cleavage site. For every cleavage site, 100 non-cleavage sites were generated, i.e., in total 83.200 non-cleavages sites (Fig. 1). The advantage of this method is that the amino acid frequencies of cleavage and non-cleavage sites remain identical. (2) All sites in the source proteins of the MHC-I ligands, that were not assigned as a cleavage site were assumed to be non-cleavage sites (N = 507.538, Fig. 1).
Prediction performance measures
Proteasome cleavage and TAP transport predictions were performed as suggested by the developers of the different prediction methods (Peters et al. 2003; Tenzer et al. 2005; Nielsen et al. 2005). The different proteasome predictors were assessed for their performance in discriminating cleavage from non-cleavage sites. First, the performance of the proteasome predictors was tested using receiver operator characteristic (ROC) curves (Swets 1988). In a ROC curve, true positive proportions (TPP) and false positive predictions (FPP) are plotted on the y- and x-axis, respectively, for every threshold. The area under the ROC curve (AUC) is a measure of the predictor’s performance. If a predictor performs well, the TPPs increase faster than the FPP, and the AUC becomes larger than 0.5; the maximal AUC is 1.0.
The AUC can only be determined on a single set of prediction scores. However, we aimed to compare the prediction performance of the proteasome predictors in combination with the TAP transport predictor. Therefore, we developed an alternative performance measure: for every TAP transport prediction value, based on the cleavage and non-cleavage sites that exceeded the TAP transport value (T), the AUC was determined ( A U C T ). If less than 25 cleavage sites or non-cleavage sites exceeded the TAP threshold, it was discarded. A score was derived by integrating over all the AUCs with respect to the TAP threshold values and subsequent normalization by the range of TAP thresholds (1). The resulting score ranges between 0 and 1, a random predictor would score 0.5 and a perfect predictor would score 1, similar to the scores obtained in an AUC analysis. This score reflects the predictive performance of the proteolysis predictor for different data sets which have been selected over a range of possible TAP values. We call this performance measure volume under the plane (VUP):
Statistics
Statistical tests were performed using the stats-package from the scipy-module in Python. The difference between AUC/VUP performance measures was determined by deriving AUCs/VUPs on 50 new data sets that were generated by bootstrapping the original data set. The derived AUCs/VUPs were compared using a paired two-tailed t test; p values less than 0.001 were considered significant (as in Tenzer et al. (2005)). We refer to this test as the ROC-comparison test.
References
Aki M, Shimbara N, Takashina M, Akiyama K, Kagawa S, Tamura T, Tanahashi N, Yoshimura T, Tanaka K, Ichihara A (1994) Interferon-gamma induces different subunit organizations and functional diversity of proteasomes. J Biochem 115(2):257–269
Clijsters L, Ogink J, Wolthuis R (2013) The spindle checkpoint, APC/CC(dc20), and APC/CC(dh1) play distinct roles in connecting mitosis to S phase. J Cell Biol 201(7):1013–1026. doi:10.1083/jcb.201211019
Emmerich NP, Nussbaum AK, Stevanovic S, Priemer M, Toes RE, Rammensee HG, Schild H (2000) The human 26 S and 20 S proteasomes generate overlapping but different sets of peptide fragments from a model protein substrate. J Biol Chem 275(28):21,140–21,148. doi:10.1074/jbc.M000740200
Florea BI, Verdoes M, Li N, van der Linden WA, Geurink PP, van den Elst H, Hofmann T, de Ru A, van Veelen PA, Tanaka K, Sasaki K, Murata S, den Dulk H, Brouwer J, Ossendorp FA, Kisselev AF, Overkleeft HS (2010) Activity-based profiling reveals reactivity of the murine thymoproteasome-specific subunit beta5t. Chem Biol 17(8):795–801. doi:10.1016/j.chembiol.2010.05.027
Gaczynska M, Rock KL, Goldberg AL (1993) Gamma-interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature 365(6443):264–267. doi:10.1038/365264a0
Geier E, Pfeifer G, Wilm M, Lucchiari-Hartz M, Baumeister W, Eichmann K, Niedermann G (1999) A giant protease with potential to substitute for some functions of the proteasome. Science 283 (5404):978–981
Ginodi I, Vider-Shalit T, Tsaban L, Louzoun Y (2008) Precise score for the prediction of peptides cleaved by the proteasome. Bioinformatics 24(4):477–483. doi:10.1093/bioinformatics/btm616,
Goldberg AL (2003) Protein degradation and protection against misfolded or damaged proteins. Nature 426(6968):895–899. doi:10.1038/nature02263
de Graaf N, van Helden MJG, Textoris-Taube K, Chiba T, Topham DJ, Kloetzel PM, Zaiss DMW, Sijts AJAM (2011) PA28 and the proteasome immunosubunits play a central and independent role in the production of MHC class I-binding peptides in vivo. Eur J Immunol 41(4):926–935. doi:10.1002/eji.201041040
Guillaume B, Chapiro J, Stroobant V, Colau D, Holle BV, Parvizi G, Bousquet-Dubouch MP, Theate I, Parmentier N, den Eynde BJV (2010) Two abundant proteasome subtypes that uniquely process some antigens presented by HLA class I molecules. Proc Natl Acad Sci USA 107(43):18,599–18,604. doi:10.1073/pnas.1009778107
Hershko A, Ciechanover A (1992) The ubiquitin system for protein degradation. Annu Rev Biochem 61:761–807. doi:10.1146/annurev.bi.61.070192.003553
Holzhutter HG, Kloetzel PM (2000) A kinetic model of vertebrate 20S proteasome accounting for the generation of major proteolytic fragments from oligomeric peptide substrates. Biophys J 79(3):1196–1205. doi:10.1016/S0006-3495(00)76374-0
Holzhutter HG, Frommel C, Kloetzel PM (1999) A theoretical approach towards the identification of cleavage-determining amino acid motifs of the 20 S proteasome. J Mol Biol 286(4):1251–1265. doi:10.1006/jmbi.1998.2530
Kesmir C, Nussbaum AK, Schild H, Detours V, Brunak S (2002) Prediction of proteasome cleavage motifs by neural networks. Protein Eng 15(4):287–296
Kesmir C, Van Noort V, De Boer RJ, Hogeweg P (2003) Bioinformatic analysis of functional differences between the immunoproteasome and the constitutive proteasome. Immunogenetics 55(7):437–449. doi:10.1007/s00251-003-0585-6
Kessler JH, Khan S, Seifert U, Gall SL, Chow KM, Paschen A, Bres-Vloemans SA, de Ru A, van Montfoort N, Franken KLMC, Benckhuijsen WE, Brooks JM, van Hall T, Ray K, Mulder A, Doxiadis, IIN, van Swieten PF, Overkleeft HS, Prat A, Tomkinson B, Neefjes J, Kloetzel PM, Rodgers DW, Hersh LB, Drijfhout JW, van Veelen PA, Ossendorp F, Melief CJM (2011) Antigen processing by nardilysin and thimet oligopeptidase generates cytotoxic T cell epitopes. Nat Immunol 12(1):45–53. doi:10.1038/ni.1974
Kincaid EZ, Che JW, York I, Escobar H, Reyes-Vargas E, Delgado JC, Welsh RM, Karow ML, Murphy AJ, Valenzuela DM, Yancopoulos GD, Rock KL (2012) Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat Immunol 13(2):129–135. doi:10.1038/ni.2203
Kloetzel PM (2001) Antigen processing by the proteasome. Nat Rev Mol Cell Biol 2(3):179–187. doi:10.1038/35056572
Kloetzel PM (2004) Generation of major histocompatibility complex class I antigens: functional interplay between proteasomes and TPPII. Nat Immunol 5(7):661–669. doi:10.1038/ni1090
Kloetzel PM, Ossendorp F (2004) Proteasome and peptidase function in MHC-class-I-mediated antigen presentation. Curr Opin Immunol 16(1):76–81
Kuttler C, Nussbaum AK, Dick TP, Rammensee HG, Schild H, Hadeler KP (2000) An algorithm for the prediction of proteasomal cleavages. J Mol Biol 298(3):417–429. doi:10.1006/jmbi.2000.3683
van Leuken R, Clijsters L, Wolthuis R (2008) To cell cycle, swing the APC/C. Biochim Biophys Acta 1786(1):49–59. doi:10.1016/j.bbcan.2008.05.002
Murata S, Sasaki K, Kishimoto T, Niwa SI, Hayashi H, Takahama Y, Tanaka K (2007) Regulation of CD8+ T cell development by thymus-specific proteasomes. Science 316(5829):1349–1353. doi:10.1126/science.1141915
Neefjes J, Jongsma MLM, Paul P, Bakke O (2011) Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol 11(12):823–836. doi:10.1038/nri3084
Nielsen M, Lundegaard C, Lund O, Kesmir C (2005) The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics 57(1-2):33–41. doi:10.1007/s00251-005-0781-7
Nussbaum AK, Kuttler C, Hadeler KP, Rammensee HG, Schild H (2001) PAProC: a prediction algorithm for proteasomal cleavages available on the WWW. Immunogenetics 53(2):87–94
Peters B, Janek K, Kuckelkorn U, Holzhutter HG (2002) Assessment of proteasomal cleavage probabilities from kinetic analysis of time-dependent product formation. J Mol Biol 318(3):847–862. doi:10.1016/S0022-2836(02)00167-5
Peters B, Bulik S, Tampe R, Endert PMV, Holzhutter HG (2003) Identifying MHC class I epitopes by predicting the TAP transport efficiency of epitope precursors. J Immunol 171(4):1741–1749
Peters J, Schönegge AM, Rockel B, Baumeister W (2011) Molecular ruler of tripeptidylpeptidase II: mechanistic principle of exopeptidase selectivity. Biochem Biophys Res Commun. 414(1):209–14. doi:10.1016/j.bbrc.2011.09.058
Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S (1999) SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50(3–4):213–219
Reits E, Griekspoor A, Neijssen J, Groothuis T, Jalink K, van Veelen P, Janssen H, Calafat J, Drijfhout JW, Neefjes J (2003) Peptide diffusion, protection, and degradation in nuclear and cytoplasmic compartments before antige presentation by MHC class I. Immunity 18(1):97–108
Saxova P, Buus S, Brunak S, Kesmir C (2003) Predicting proteasomal cleavage sites: a comparison of available methods. Int Immunol 15(7):781–787
Seifert U, Maranon C, Shmueli A, Desoutter JF, Wesoloski L, Janek K, Henklein P, Diescher S, Andrieu M, de la Salle H, Weinschenk T, Schild H, Laderach D, Galy A, Haas G, Kloetzel PM, Reiss Y, Hosmalin A (2003) An essential role for tripeptidyl peptidase in the generation of an MHC class I epitope. Nat Immunol 4(4):375–379. doi:10.1038/ni905
Seifert U, Bialy LP, Ebstein F, Bech-Otschir D, Voigt A, Schroter F, Prozorovski T, Lange N, Steffen J, Rieger M, Kuckelkorn U, Aktas O, Kloetzel PM, Kruger E (2010) Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell 142(4):613–624. doi:10.1016/j.cell.2010.07.036
Shen XZ, Lukacher AE, Billet S, Williams IR, Bernstein KE (2008) Expression of angiotensin-converting enzyme changes major histocompatibility complex class I peptide presentation by modifying C termini of peptide precursors. J Biol Chem 283(15):9957–9965. doi:10.1074/jbc.M709574200
Shen XZ, Billet S, Lin C, Okwan-Duodu D, Chen X, Lukacher AE, Bernstein KE (2011) The carboxypeptidase ACE shapes the MHC class I peptide repertoire. Nat Immunol 12(11):1078–1085. doi:10.1038/ni.2107
Swets JA (1988) Measuring the accuracy of diagnostic systems. Science 240(4857):1285–1293
Tenzer S, Stoltze L, Schonfisch B, Dengjel J, Muller M, Stevanovic S, Rammensee HG, Schild H (2004) Quantitative analysis of prion-protein degradation by constitutive and immuno-20S proteasomes indicates differences correlated with disease susceptibility. J Immunol 172(2):1083–1091
Tenzer S, Peters B, Bulik S, Schoor O, Lemmel C, Schatz MM, Kloetzel PM, Rammensee HG, Schild H, Holzhutter HG (2005) Modeling the MHC class I pathway by combining predictions of proteasomal cleavage, TAP transport and MHC class I binding. Cell Mol Life Sci 62(9):1025–1037
Toes RE, Nussbaum AK, Degermann S, Schirle M, Emmerich NP, Kraft M, Laplace C, Zwinderman A, Dick TP, Muller J, Schonfisch B, Schmid C, Fehling HJ, Stevanovic S, Rammensee HG, Schild H (2001) Discrete cleavage motifs of constitutive and immunoproteasomes revealed by quantitative analysis of cleavage products. J Exp Med 194(1):1–12
Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, Salimi N, Damle R, Sette A, Peters B (2010) The immune epitope database 2.0. Nucleic Acids Res 38(Database issue):D854–D862. doi:10.1093/nar/gkp1004
Acknowledgments
We thank Morten Nielsen, Becca Asquith, Bjoern Peters, Berend Snel, Ilka Hoof, Hanneke van Deutekom, and Xiangyu Rao for discussion on this research project and technical support. This study was financially supported by the Netherlands Organization for Scientific Research (www.nwo.nl, Computational Life Sciences Program, grant number 635.100.025), and by Utrecht University ( www.uu.nl). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. S1
Predicting in vivo proteolysis by combining proteolysis and TAP transport predictor scores, with the alternative non-cleavage site definition. For the different proteasome cleavage predictors, the proteasome cleavage prediction score was added to the TAP transport prediction score (as proposed by Tenzer et al. 2005). Prediction performance was measured as the AUC of an ROC-curve (Y-axis), using all other sites in a source proteins as non-cleavage sites (see Methods). When combining the scores, the weight of the TAP transport score was changed by the factor W (on the X-axis). The combined score (C), based on the TAP transport (T) and proteolysis (P) score would than be C = W * T + P. As a result, the proteasome cleavage or the TAP transport predictor has a larger influence in the combined score if W is smaller or larger, respectively. See Figure 3 for color coding. (PDF 34.8 KB)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits use, duplication, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Calis, J.J.A., Reinink, P., Keller, C. et al. Role of peptide processing predictions in T cell epitope identification: contribution of different prediction programs. Immunogenetics 67, 85–93 (2015). https://doi.org/10.1007/s00251-014-0815-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-014-0815-0