Abstract
Interest in research into bioactive peptides (BPs) is growing because of their health-promoting ability. Several bioactivities have been ascribed to peptides, including antioxidant, antihypertensive and antimicrobial properties. As they can be produced from precursor proteins, the investigation of BPs in foods is becoming increasingly popular. For the same reason, production of BPs from by-products has also emerged as a possible means of reducing waste and recovering value-added compounds suitable for functional food production and supplements. Milk, meat and fish are the most investigated sources of BPs, but vegetable-derived peptides are also of interest. Vegetables are commonly consumed, and agro-industrial wastes constitute a cheap, large and lower environmental impact source of proteins. The use of advanced analytical techniques for separation and identification of peptides would greatly benefit the discovery of new BPs. In this context, this review provides an overview of the most recent applications in BP investigations for vegetable food and by-products. The most important issues regarding peptide isolation and separation, by single or multiple chromatographic techniques, are discussed. Additionally, problems connected with peptide identification in plants and non-model plants are discussed regarding the particular case of BP identification. Finally, the issue of peptide validation to confirm sequence and bioactivity is presented.

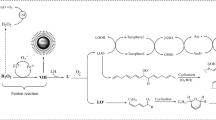
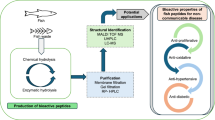
Graphical representation of the analytical workflow needed for investigation of bioactive peptides and applied to vegetables and vegetable wastes
Graphical Abstract
Similar content being viewed by others
References
Zenezini Chiozzi R, Capriotti AL, Cavaliere C, La Barbera G, Piovesana S, Samperi R, et al. Purification and identification of endogenous antioxidant and ACE-inhibitory peptides from donkey milk by multidimensional liquid chromatography and nanoHPLC-high resolution mass spectrometry. Anal Bioanal Chem. 2016;408:5657–66. https://doi.org/10.1007/s00216-016-9672-z.
Rizzello CG, Tagliazucchi D, Babini E, Sefora Rutella G, Taneyo Saa DL, Gianotti A. Bioactive peptides from vegetable food matrices: research trends and novel biotechnologies for synthesis and recovery. J Funct Foods. 2016;27:549–69. https://doi.org/10.1016/j.jff.2016.09.023.
Samperi R, Capriotti AL, Cavaliere C, Colapicchioni V, Chiozzi RZ, Laganà A. Food proteins and peptides. In: Barcelo D, editor. Comprehensive Analytical Chemistry. vol 68. Amsterdam: Elsevier; 2015. pp 309–357. https://doi.org/10.1016/B978-0-444-63340-8.00006-6
Hettiarachchy NS. Bioactive food proteins and peptides: applications in human health. Boca Raton: CRC Press; 2012.
Capriotti AL, Cavaliere C, Piovesana S, Samperi R, Laganà A. Recent trends in the analysis of bioactive peptides in milk and dairy products. Anal Bioanal Chem. 2016;408:2677–85. https://doi.org/10.1007/s00216-016-9303-8.
Piovesana S, Capriotti AL, Cavaliere C, La Barbera G, Samperi R, Zenezini Chiozzi R, et al. Peptidome characterization and bioactivity analysis of donkey milk. J Proteome. 2015;119:21–9. https://doi.org/10.1016/j.jprot.2015.01.020.
Yu Z, Yin Y, Zhao W, Chen F, Liu J. Application and bioactive properties of proteins and peptides derived from hen eggs: opportunities and challenges. J Sci Food Agric. 2014;94:2839–45. https://doi.org/10.1002/jsfa.6670.
Halim NRA, Yusof HM, Sarbon NM. Functional and bioactive properties of fish protein hydolysates and peptides: a comprehensive review. Trends Food Sci Technol. 2016;51:24–33. https://doi.org/10.1016/j.tifs.2016.02.007.
Lafarga T, Hayes M. Bioactive peptides from meat muscle and by-products: generation, functionality and application as functional ingredients. Meat Sci. 2014;98:227–39. https://doi.org/10.1016/j.meatsci.2014.05.036.
Daskaya-Dikmen C, Yucetepe A, Karbancioglu-Guler F, Daskaya H, Ozcelik B. Angiotensin-I-converting enzyme (ACE)-inhibitory peptides from plants. Nutrients. 2017;9:1–19. https://doi.org/10.3390/nu9040316.
La Barbera G, Capriotti AL, Cavaliere C, Montone CM, Piovesana S, Samperi R, et al. Liquid chromatography-high resolution mass spectrometry for the analysis of phytochemicals in vegetal-derived food and beverages. Food Res Int. 2017;100:28–52. https://doi.org/10.1016/j.foodres.2017.07.080.
Meneguetti BT, Machado L dos S, Oshiro KGN, Nogueira ML, Carvalho CME, Franco OL. Antimicrobial peptides from fruits and their potential use as biotechnological tools—a review and outlook. Front Microbiol. 2017;7:2136. https://doi.org/10.3389/fmicb.2016.02136.
Hartmann R, Meisel H. Food-derived peptides with biological activity: from research to food applications. Curr Opin Biotechnol. 2007;18:163–9. https://doi.org/10.1016/j.copbio.2007.01.013.
García MC, Puchalska P, Esteve C, Marina ML. Vegetable foods: a cheap source of proteins and peptides with antihypertensive, antioxidant, and other less occurrence bioactivities. Talanta. 2013;106:328–49. https://doi.org/10.1016/j.talanta.2012.12.041.
Lee SY, Hur SJ. Antihypertensive peptides from animal products, marine organisms, and plants. Food Chem. 2017;228:506–17. https://doi.org/10.1016/j.foodchem.2017.02.039.
Mine Y, Li-Chan E, Jiang B, editors. Bioactive proteins and peptides as functional foods and nutraceuticals. Oxford: Wiley-Blackwell; 2010. https://doi.org/10.1002/9780813811048.
Chen M, Li B. The effect of molecular weights on the survivability of casein-derived antioxidant peptides after the simulated gastrointestinal digestion. Innovative Food Sci Emerg Technol. 2012;16:341–8. https://doi.org/10.1016/j.ifset.2012.07.009.
Torres-Fuentes C, Contreras MDM, Recio I, Alaiz M, Vioque J. Identification and characterization of antioxidant peptides from chickpea protein hydrolysates. Food Chem. 2015;180:194–202. https://doi.org/10.1016/j.foodchem.2015.02.046.
Zou TB, He TP, Li, HB, Tang HW, Xia EQ (2016) The structure-activity relationship of the antioxidant peptides from natural proteins. Molecules. https://doi.org/10.3390/molecules21010072.
Tovar-Pérez EG, Guerrero-Becerra L, Lugo-Cervantes E. Antioxidant activity of hydrolysates and peptide fractions of glutelin from cocoa ( Theobroma cacao L.) seed. CyTA J Food. 2017;15:489–96. https://doi.org/10.1080/19476337.2017.1297963.
Word Health Organization. Global status report on noncommunicable diseases 2014. Geneva: World Health Organization; 2014.
Dhaval A, Yadav N, Purwar S. Potential applications of food derived bioactive peptides in management of health. Int J Pept Res Ther. 2016;22:377–98. https://doi.org/10.1007/s10989-016-9514-z.
Visvanathan R, Jayathilake C, Chaminda Jayawardana B, Liyanage R. Health-beneficial properties of potato and compounds of interest. J Sci Food Agric. 2016;96:4850–60. https://doi.org/10.1002/jsfa.7848.
Rudolph S, Lunow D, Kaiser S, Henle T. Identification and quantification of ACE-inhibiting peptides in enzymatic hydrolysates of plant proteins. Food Chem. 2017;224:19–25. https://doi.org/10.1016/j.foodchem.2016.12.039.
Zhang M, Mu T-H. Identification and characterization of antioxidant peptides from sweet potato protein hydrolysates by Alcalase under high hydrostatic pressure. Innovative Food Sci Emerg Technol. 2017;43:92–101. https://doi.org/10.1016/j.ifset.2017.08.001.
Babini E, Tagliazucchi D, Martini S, Dei Più L, Gianotti A. LC-ESI-QTOF-MS identification of novel antioxidant peptides obtained by enzymatic and microbial hydrolysis of vegetable proteins. Food Chem. 2017;228:186–96. https://doi.org/10.1016/j.foodchem.2017.01.143.
Agrawal H, Joshi R, Gupta M. Isolation and characterisation of enzymatic hydrolysed peptides with antioxidant activities from green tender sorghum. LWT Food Sci Technol. 2017;84:608–16. https://doi.org/10.1016/j.lwt.2017.06.036.
Agrawal H, Joshi R, Gupta M. Isolation, purification and characterization of antioxidative peptide of pearl millet (Pennisetum glaucum) protein hydrolysate. Food Chem. 2016;204:365–72. https://doi.org/10.1016/j.foodchem.2016.02.127.
Yan QJ, Huang LH, Sun Q, Jiang ZQ, Wu X. Isolation, identification and synthesis of four novel antioxidant peptides from rice residue protein hydrolyzed by multiple proteases. Food Chem. 2015;179:290–5. https://doi.org/10.1016/j.foodchem.2015.01.137.
Ren G, Zhu Y, Shi Z. Detection of lunasin in quinoa (Chenopodium quinoa, Willd) and the in vitro evaluation of its antioxidant and anti-inflammatory activities. J Sci Food Agric. 2017;28:303–25. https://doi.org/10.1002/jsfa.8278.
Sabbione AC, Ibañez SM, Martínez EN, Añón MC, Scilingo AA. Antithrombotic and antioxidant activity of amaranth hydrolysate obtained by activation of an endogenous protease. Plant Foods Hum Nutr. 2016;71:174–82. https://doi.org/10.1007/s11130-016-0540-y.
Jamdar SN, Deshpande R, Marathe SA. Effect of processing conditions and in vitro protein digestion on bioactive potentials of commonly consumed legumes. Food Biosci. 2017;20:1–11. https://doi.org/10.1016/j.fbio.2017.07.007.
Garcia-Mora P, Peñas E, Frias J, Gomez R, Martinez-Villaluenga C. High-pressure improves enzymatic proteolysis and the release of peptides with angiotensin I converting enzyme inhibitory and antioxidant activities from lentil proteins. Food Chem. 2015;171:224–32. https://doi.org/10.1016/j.foodchem.2014.08.116.
Singh BP, Vij S. Growth and bioactive peptides production potential of Lactobacillus plantarum strain C2 in soy milk: a LC-MS/MS based revelation for peptides biofunctionality. LWT Food Sci Technol. 2017;86:293–301. https://doi.org/10.1016/j.lwt.2017.08.013.
Vallabha VS, Tiku PK. Antihypertensive peptides derived from soy protein by fermentation. Int J Pept Res Ther. 2014;20:161–8. https://doi.org/10.1007/s10989-013-9377-5.
Gu Y, Wu J. LC-MS/MS coupled with QSAR modeling in characterising of angiotensin I-converting enzyme inhibitory peptides from soybean proteins. Food Chem. 2013;141:2682–90. https://doi.org/10.1016/j.foodchem.2013.04.064.
Sornwatana T, Bangphoomi K, Roytrakul S, Wetprasit N, Choowongkomon K, Ratanapo S. Chebulin: Terminalia chebula Retz. fruit-derived peptide with angiotensin-I-converting enzyme inhibitory activity. Biotechnol Appl Biochem. 2015;62:746–53. https://doi.org/10.1002/bab.1321.
Liu M, Du M, Zhang Y, Xu W, Wang C, Wang K, et al. Purification and identification of an ACE inhibitory peptide from walnut protein. J Agric Food Chem. 2013;61:4097–100. https://doi.org/10.1021/jf4001378.
Vilcacundo R, Martínez-Villaluenga C, Hernández-Ledesma B. Release of dipeptidyl peptidase IV, α-amylase and α-glucosidase inhibitory peptides from quinoa (Chenopodium quinoa Willd.) during in vitro simulated gastrointestinal digestion. J Funct Foods. 2017;35:531–9. https://doi.org/10.1016/j.jff.2017.06.024.
Jakubczyk A, Karaś M, Złotek U, Szymanowska U. Identification of potential inhibitory peptides of enzymes involved in the metabolic syndrome obtained by simulated gastrointestinal digestion of fermented bean (Phaseolus vulgaris L.) seeds. Food Res Int. 2017;100:489–96. https://doi.org/10.1016/j.foodres.2017.07.046.
Mäkinen S, Streng T, Larsen LB, Laine A, Pihlanto A. Angiotensin I-converting enzyme inhibitory and antihypertensive properties of potato and rapeseed protein-derived peptides. J Funct Foods. 2016;25:160–73. https://doi.org/10.1016/j.jff.2016.05.016.
Soares RA, Mendonça S, de Castro L, Menezes AC, Arêas JA. Major peptides from amaranth (Amaranthus cruentus) protein inhibit HMG-CoA reductase activity. Int J Mol Sci. 2015;16:4150–60. https://doi.org/10.3390/ijms16024150.
Capriotti AL, Caruso G, Cavaliere C, Samperi R, Ventura S, Zenezini Chiozzi R, et al. Identification of potential bioactive peptides generated by simulated gastrointestinal digestion of soybean seeds and soy milk proteins. J Food Compos Anal. 2015;44:205–13. https://doi.org/10.1016/j.jfca.2015.08.007.
Dia VP, Krishnan HB. BG-4, a novel anticancer peptide from bitter gourd (Momordica charantia), promotes apoptosis in human colon cancer cells. Sci Rep. 2016;6:33532. https://doi.org/10.1038/srep33532.
Rayaprolu SJ, Hettiarachchy NS, Horax R, Kumar-Phillips G, Liyanage R, Lay J, et al. Purification and characterization of a peptide from soybean with cancer cell proliferation inhibition. J Food Biochem. 2017;41:e12374. https://doi.org/10.1111/jfbc.12374.
Wang X, Chen H, Fu X, Li S, Wei J. A novel antioxidant and ACE inhibitory peptide from rice bran protein: biochemical characterization and molecular docking study. LWT Food Sci Technol. 2017;75:93–9. https://doi.org/10.1016/j.lwt.2016.08.047.
White BL, Sanders TH, Davis JP. Potential ACE-inhibitory activity and nanoLC-MS/MS sequencing of peptides derived from aflatoxin contaminated peanut meal. LWT Food Sci Technol. 2014;56:537–42. https://doi.org/10.1016/j.lwt.2013.11.039.
Connolly A, O’Keeffe MB, Piggott CO, Nongonierma AB, Fitzgerald RJ. Generation and identification of angiotensin converting enzyme (ACE) inhibitory peptides from a brewers’ spent grain protein isolate. Food Chem. 2015;176:64–71. https://doi.org/10.1016/j.foodchem.2014.12.027.
García MC, Endermann J, González-García E, Marina ML. HPLC-Q-TOF-MS Identification of antioxidant and antihypertensive peptides recovered from cherry (Prunus cerasus L.) subproducts. J Agric Food Chem. 2015;63:1514–20. https://doi.org/10.1021/jf505037p.
Vásquez-Villanueva R, Marina ML, García MC. Identification by hydrophilic interaction and reversed-phase liquid chromatography-tandem mass spectrometry of peptides with antioxidant capacity in food residues. J Chromatogr A. 2016;1428:185–92. https://doi.org/10.1016/j.chroma.2015.07.032.
Zenezini Chiozzi R, Capriotti AL, Cavaliere C, La Barbera G, Piovesana S, Laganà A. Identification of three novel angiotensin-converting enzyme inhibitory peptides derived from cauliflower by-products by multidimensional liquid chromatography and bioinformatics. J Funct Foods. 2016;27:262–73. https://doi.org/10.1016/j.jff.2016.09.010.
Furuta T, Miyabe Y, Yasui H, Kinoshita Y, Kishimura H. Angiotensin I converting enzyme inhibitory peptides derived from phycobiliproteins of dulse Palmaria palmata. Mar Drugs. 2016;14:32–42. https://doi.org/10.3390/md14020032.
Harnedy PA, O’Keeffe MB, Fitzgerald RJ. Purification and identification of dipeptidyl peptidase (DPP) IV inhibitory peptides from the macroalga Palmaria palmata. Food Chem. 2015;172:400–6. https://doi.org/10.1016/j.foodchem.2014.09.083.
Moayedi A, Mora L, Aristoy MC, Hashemi M, Safari M, Toldrá F. ACE-inhibitory and antioxidant activities of peptide fragments obtained from tomato processing by-products fermented using Bacillus subtilis: effect of amino acid composition and peptides molecular mass distribution. Appl Biochem Biotechnol. 2017;181:48–64. https://doi.org/10.1007/s12010-016-2198-1.
Waglay A, Karboune S. Enzymatic generation of peptides from potato proteins by selected proteases and characterization of their structural properties. Biotechnol Prog. 2016;32:420–9. https://doi.org/10.1002/btpr.2245.
Xie N, Huang J, Li B, Cheng J, Wang Z, Yin J, et al. Affinity purification and characterisation of zinc chelating peptides from rapeseed protein hydrolysates: possible contribution of characteristic amino acid residues. Food Chem. 2015;173:210–7. https://doi.org/10.1016/j.foodchem.2014.10.030.
Ambigaipalan P, Al-Khalifa AS, Shahidi F. Antioxidant and angiotensin I converting enzyme (ACE) inhibitory activities of date seed protein hydrolysates prepared using Alcalase, Flavourzyme and thermolysin. J Funct Foods. 2015;18:1125–37. https://doi.org/10.1016/j.jff.2015.01.021.
Ghorab H, Lammi C, Arnoldi A, Kabouche Z, Aiello G. Proteomic analysis of sweet algerian apricot kernels (Prunus armeniaca L.) by combinatorial peptide ligand libraries and LC–MS/MS. Food Chem. 2018;239:935–45. https://doi.org/10.1016/j.foodchem.2017.07.054.
Stefanucci A, Mollica A, Macedonio G, Zengin G, Ahmed AA, Novellino E. Exogenous opioid peptides derived from food proteins and their possible uses as dietary supplements: a critical review. Food Rev Int. 2018;34:70–86. https://doi.org/10.1080/87559129.2016.1225220.
Cao Y, Miao J, Liu G, Luo Z, Xia Z, Liu F, et al. Bioactive peptides isolated from casein phosphopeptides enhance calcium and magnesium uptake in Caco-2 cell monolayers. J Agric Food Chem. 2017;65:2307–14. https://doi.org/10.1021/acs.jafc.6b05711.
Meisel H, RJ FG. Biofunctional peptides from milk proteins: mineral binding and cytomodulatory effects. Curr Pharm Des. 2003;9:1289–95. https://doi.org/10.2174/1381612033454847.
Ebner J, Aşçi Arslan A, Fedorova M, Hoffmann R, Küçükçetin A, Pischetsrieder M. Peptide profiling of bovine kefir reveals 236 unique peptides released from caseins during its production by starter culture or kefir grains. J Proteome. 2015;117:41–57. https://doi.org/10.1016/j.jprot.2015.01.005.
Lv Y, Bao XL, Yang BC, Ren CG, Guo ST (2008) Effect of soluble soybean protein hydrolysate-calcium complexes on calcium uptake by Caco-2 cells. J Food Sci. https://doi.org/10.1111/j.1750-3841.2008.00873.x.
Anjum K, Abbas SQ, Akhter N, Shagufta BI, Shah SAA, ul HSS. Emerging biopharmaceuticals from bioactive peptides derived from marine organisms. Chem Biol Drug Des. 2017;49:34–47. https://doi.org/10.1111/cbdd.12925.
Aziz M, Karboune S (2017) Natural antimicrobial/antioxidant agents in meat and poultry products as well as fruits and vegetables: a review. Crit Rev Food Sci Nutr. https://doi.org/10.1080/10408398.2016.1194256.
Mulder KCL, Lima LA, Miranda VJ, Dias SC, Franco OL. Current scenario of peptide-based drugs: the key roles of cationic antitumor and antiviral peptides. Front Microbiol. 2013;4:321. https://doi.org/10.3389/fmicb.2013.00321.
Zhou X, Wen L, Li Z, Zhou Y, Chen Y, Lu Y. Advance on the benefits of bioactive peptides from buckwheat. Phytochem Rev. 2015;14:381–8. https://doi.org/10.1007/s11101-014-9390-0.
Agyei D. Bioactive proteins and peptides from soybeans. Recent Pat Food Nutr Agric. 2015;7:100–7. https://doi.org/10.2174/2212798407666150629134141.
Ramdath D, Padhi E, Sarfaraz S, Renwick S, Duncan A. Beyond the cholesterol-lowering effect of soy protein: a review of the effects of dietary soy and its constituents on risk factors for cardiovascular disease. Nutrients. 2017;9:324. https://doi.org/10.3390/NU9040324.
Lovati MR, Manzoni C, Gianazza E, Arnoldi A, Kurowska E, Carroll KK, et al. Soy protein peptides regulate cholesterol homeostasis in Hep G2 cells. J Nutr. 2000;130:2543–9.
Cho SJ, Juillerat MA, Lee CH. Identification of LDL-receptor transcription stimulating peptides from soybean hydrolysate in human hepatocytes. J Agric Food Chem. 2008;56:4372–6. https://doi.org/10.1021/jf800676a.
Mochizuki Y, Maebuchi M, Kohno M, Hirotsuka M, Wadahama H, Moriyama T, et al. Changes in lipid metabolism by soy β-conglycinin-derived peptides in HepG2 cells. J Agric Food Chem. 2009;57:1473–80. https://doi.org/10.1021/jf8031793.
Liu Y, Yang J, Lei L, Wang L, Wang X, Ying Ma K, et al. 7S protein is more effective than total soybean protein isolate in reducing plasma cholesterol. J Funct Foods. 2017;36:18–26. https://doi.org/10.1016/j.jff.2017.06.039.
Wang W, Gonzalez De Mejia E. A new frontier in soy bioactive peptides that may prevent age-related chronic diseases. Compr Rev Food Sci Food Saf. 2005;4:63–78. https://doi.org/10.1111/j.1541-4337.2005.tb00075.x.
Omoni AO, Aluko RE. Effect of cationic flaxseed protein hydrolysate fractions on the in vitro structure and activity of calmodulin-dependent endothelial nitric oxide synthase. Mol Nutr Food Res. 2006;50:958–66. https://doi.org/10.1002/mnfr.200600041.
Li H, Aluko RE. Structural modulation of calmodulin and calmodulin-dependent protein kinase II by pea protein hydrolysates. Int J Food Sci Nutr. 2006;57:178–89. https://doi.org/10.1080/09637480600659144.
da Silva Dantas CC, de Souza EL, Cardoso JD, de Lima LA, de Sousa Oliveira K, Migliolo L, et al. Identification of a napin-like peptide from Eugenia malaccensis L. seeds with inhibitory activity toward Staphylococcus aureus and Salmonella Enteritidis. Protein J. 2014;33:549–56. https://doi.org/10.1007/s10930-014-9587-5.
Fesenko IA, Arapidi GP, Skripnikov A, Alexeev DG, Kostryukova ES, Manolov AI, et al. Specific pools of endogenous peptides are present in gametophore, protonema, and protoplast cells of the moss Physcomitrella patens. BMC Plant Biol. 2015;15:87. https://doi.org/10.1186/s12870-015-0468-7.
Minkiewicz P, Dziuba J, Iwaniak A, Dziuba M, Darewicz M. BIOPEP database and other programs for processing bioactive peptide sequences. J AOAC Int. 2008;91:965–80.
Ahmadifard N, Murueta JHC, Abedian-Kenari A, Motamedzadegan A, Jamali H. Comparison the effect of three commercial enzymes for enzymatic hydrolysis of two substrates (rice bran protein concentrate and soy-been protein) with SDS-PAGE. J Food Sci Technol. 2016;53:1279–84. https://doi.org/10.1007/s13197-015-2087-6.
Hoppe A, Jung S, Patnaik A, Zeece MG. Effect of high pressure treatment on egg white protein digestibility and peptide products. Innovative Food Sci Emerg Technol. 2013;17:54–62. https://doi.org/10.1016/j.ifset.2012.11.003.
López-Expósito I, Chicón R, Belloque J, Recio I, Alonso E, López-Fandiño R. Changes in the ovalbumin proteolysis profile by high pressure and its effect on IgG and IgE binding. J Agric Food Chem. 2008;56:11809–16. https://doi.org/10.1021/jf8023613.
Zhang T, Jiang B, Miao M, Mu W, Li Y. Combined effects of high-pressure and enzymatic treatments on the hydrolysis of chickpea protein isolates and antioxidant activity of the hydrolysates. Food Chem. 2012;135:904–12. https://doi.org/10.1016/j.foodchem.2012.05.097.
Li G-H, Qu M-R, Wan J-Z, You J-M. Antihypertensive effect of rice protein hydrolysate with in vitro angiotensin I-converting enzyme inhibitory activity in spontaneously hypertensive rats. Asia Pac J Clin Nutr. 2007;16 Suppl 1:275–80. https://doi.org/10.6133/apjcn.2007.16.s1.52.
Rayaprolu SJ, Hettiarachchy NS, Chen P, Kannan A, Mauromostakos A. Peptides derived from high oleic acid soybean meals inhibit colon, liver and lung cancer cell growth. Food Res Int. 2013;50:282–8. https://doi.org/10.1016/j.foodres.2012.10.021.
Esteve C, Marina ML, García MC. Novel strategy for the revalorization of olive (Olea europaea) residues based on the extraction of bioactive peptides. Food Chem. 2015;167:272–80. https://doi.org/10.1016/j.foodchem.2014.06.090.
Lemes AC, Sala L, Ores JDC, Braga ARC, Egea MB, Fernandes KF. A review of the latest advances in encrypted bioactive peptides from protein-rich waste. Int J Mol Sci. 2016;17:950. https://doi.org/10.3390/ijms17060950.
Banerjee J, Singh R, Vijayaraghavan R, MacFarlane D, Patti AF, Arora A. Bioactives from fruit processing wastes: green approaches to valuable chemicals. Food Chem. 2017;225:10–22. https://doi.org/10.1016/j.foodchem.2016.12.093.
Capriotti AL, Caruso G, Cavaliere C, Piovesana S, Samperi R, Laganà A. Comparison of three different enrichment strategies for serum low molecular weight protein identification using shotgun proteomics approach. Anal Chim Acta. 2012;740:58–65. https://doi.org/10.1016/j.aca.2012.06.033.
Capriotti AL, Cavaliere C, Foglia P, Piovesana S, Samperi R, Zenezini Chiozzi R, et al. Development of an analytical strategy for the identification of potential bioactive peptides generated by in vitro tryptic digestion of fish muscle proteins. Anal Bioanal Chem. 2015;407:845–54. https://doi.org/10.1007/s00216-014-8094-z.
Guijarro-Díez M, García MC, Crego AL, Marina ML. Off-line two dimensional isoelectrofocusing-liquid chromatography/mass spectrometry (time of flight) for the determination of the bioactive peptide lunasin. J Chromatogr A. 2014;1371:117–24. https://doi.org/10.1016/j.chroma.2014.10.019.
Capriotti AL, Cavaliere C, Cavazzini A, Gasparrini F, Pierri G, Piovesana S, et al. A multidimensional liquid chromatography–tandem mass spectrometry platform to improve protein identification in high-throughput shotgun proteomics. J Chromatogr A. 2017;1498:176–82. https://doi.org/10.1016/j.chroma.2017.03.032.
Ruprecht B, Wang D, Zenezini Chiozzi R, Li LH, Hahne H, Kuster B. Hydrophilic strong anion exchange (hSAX) chromatography enables deep fractionation of tissue proteomes. Methods Mol Biol. 2017;1550:69–82. https://doi.org/10.1007/978-1-4939-6747-6_7.
Štěpánová S, Kašička V. Analysis of proteins and peptides by electromigration methods in microchips. J Sep Sci. 2017;40:228–50. https://doi.org/10.1002/jssc.201600962.
Heemskerk AAM, Deelder AM, Mayboroda OA. CE-ESI-MS for bottom-up proteomics: advances in separation, interfacing and applications. Mass Spectrom Rev. 2016;35:259–71. https://doi.org/10.1002/mas.21432.
Sun L, Zhu G, Yan X, Zhang Z, Wojcik R, Champion MM, et al. Capillary zone electrophoresis for bottom-up analysis of complex proteomes. Proteomics. 2016;16:188–96. https://doi.org/10.1002/pmic.201500339.
Mitulović G. New HPLC techniques for proteomics analysis: a short overview of latest developments. J Liq Chromatogr Relat Technol. 2015;38:390–403. https://doi.org/10.1080/10826076.2014.941266.
Gilar M, Olivova P, Daly AE, Gebler JC. Orthogonality of separation in two-dimensional liquid chromatography. Anal Chem. 2005;77:6426–34. https://doi.org/10.1021/ac050923i.
Zenezini Chiozzi R, Capriotti AL, Cavaliere C, La Barbera G, Montone CM, Piovesana S, Laganà A (2017) Label-free shotgun proteomics approach to characterize muscle tissue from farmed and wild European sea bass (Dicentrarchus labrax). Food Anal Methods. https://doi.org/10.1007/s12161-017-0999-7.
Capriotti AL, Caruso G, Cavaliere C, Samperi R, Stampachiacchiere S, Zenezini Chiozzi R, et al. Protein profile of mature soybean seeds and prepared soybean milk. J Agric Food Chem. 2014;62:9893–9. https://doi.org/10.1021/jf5034152.
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;465:120. https://doi.org/10.1038/nature08957.
Buszewski B, Noga S. Hydrophilic interaction liquid chromatography (HILIC)-a powerful separation technique. Anal Bioanal Chem. 2012;402:231–47. https://doi.org/10.1007/s00216-011-5308-5.
Boudesocque L, Lameiras P, Amarouche N, Giraud M, Quattrini F, Garrity JM, et al. Ion-exchange centrifugal partition chromatography: a methodological approach for peptide separation. J Chromatogr A. 2012;1236:115–22. https://doi.org/10.1016/j.chroma.2012.03.010.
Boudesocque L, Kapel R, Paris C, Dhulster P, Marc I, Renault JH. Concentration and selective fractionation of an antihypertensive peptide from an alfalfa white proteins hydrolysate by mixed ion-exchange centrifugal partition chromatography. J Chromatogr B Anal Technol Biomed Life Sci. 2012;905:23–30. https://doi.org/10.1016/j.jchromb.2012.07.034.
Domínguez-Vega E, Kotkowska O, Concepción García M, Crego AL, Marina ML. Fast determination of the functional peptide soymetide in different soybean derived foods by capillary-high performance liquid chromatography. J Chromatogr A. 2011;1218:4928–33. https://doi.org/10.1016/j.chroma.2011.05.055.
Zenezini Chiozzi R, Capriotti AL, Cavaliere C, Ferraris F, La Barbera G, Piovesana S, et al. Evaluation of column length and particle size effect on the untargeted profiling of a phytochemical mixture by using UHPLC coupled to high-resolution mass spectrometry. J Sep Sci. 2017;40:2541–255. https://doi.org/10.1002/jssc.201700135.
Bazinet L, Firdaous L. Separation of bioactive peptides by membrane processes: technologies and devices. Recent Pat Biotechnol. 2013;7:9–27. https://doi.org/10.2174/1872208311307010003.
Berg JM, Tymoczko JL, Stryer L. Biochemistry. 5th ed. New York: Freeman; 2006.
Siow HL, Gan CY. Extraction of antioxidative and antihypertensive bioactive peptides from Parkia speciosa seeds. Food Chem. 2013;141:3435–42. https://doi.org/10.1016/j.foodchem.2013.06.030.
Capriotti AL, Cavaliere C, Foglia P, Piovesana S, Samperi R, Stampachiacchiere S, et al. Proteomic platform for the identification of proteins in olive (Olea europaea) pulp. Anal Chim Acta. 2013;800:36–42. https://doi.org/10.1016/j.aca.2013.09.014.
Capriotti AL, Caruso G, Cavaliere C, Foglia P, Piovesana S, Samperi R, et al. Proteome investigation of the non-model plant pomegranate (Punica granatum L.). Anal Bioanal Chem. 2013;405:9301–9. https://doi.org/10.1007/s00216-013-7382-3.
Mohayeji M, Capriotti AL, Cavaliere C, Piovesana S, Samperi R, Stampachiacchiere S, et al. Heterosis profile of sunflower leaves: a label free proteomics approach. J Proteome. 2014;99:101–10. https://doi.org/10.1016/j.jprot.2014.01.028.
Capriotti AL, Cavaliere C, Piovesana S, Stampachiacchiere S, Ventura S, Zenezini Chiozzi R, et al. Characterization of quinoa seed proteome combining different protein precipitation techniques: improvement of knowledge of nonmodel plant proteomics. J Sep Sci. 2015;38:1017–25. https://doi.org/10.1002/jssc.201401319.
Armengaud J, Trapp J, Pible O, Geffard O, Chaumot A, Hartmann EM. Non-model organisms, a species endangered by proteogenomics. J Proteome. 2014;105:5–18. https://doi.org/10.1016/j.jprot.2014.01.007.
Yan Y, Kusalik AJ, Wu FX. NovoExD: de novo peptide sequencing for ETD/ECD spectra. IEEE/ACM Trans Comput Biol Bioinforma. 2017;14:337–44. https://doi.org/10.1109/TCBB.2015.2389813.
Ye X, Zhao N, Yu X, Han X, Gao H, Zhang X. Extensive characterization of peptides from Panax ginseng C. A. Meyer using mass spectrometric approach. Proteomics. 2016;16:2788–91. https://doi.org/10.1002/pmic.201600183.
Udenigwe CC. Bioinformatics approaches, prospects and challenges of food bioactive peptide research. Trends Food Sci Technol. 2014;36:137–43. https://doi.org/10.1016/j.tifs.2014.02.004.
Li-Chan ECY. Bioactive peptides and protein hydrolysates: research trends and challenges for application as nutraceuticals and functional food ingredients. Curr Opin Food Sci. 2015;1:28–37. https://doi.org/10.1016/j.cofs.2014.09.005.
Wang T-Y, Hsieh C-H, Hung C-C, Jao C-L, Lin P-Y, Hsieh Y-L, et al. A study to evaluate the potential of an in silico approach for predicting dipeptidyl peptidase-IV inhibitory activity in vitro of protein hydrolysates. Food Chem. 2017;234:431–8. https://doi.org/10.1016/j.foodchem.2017.05.035.
Cherkasov A, Muratov EN, Fourches D, Varnek A, Baskin II, Cronin M, et al. QSAR modeling: Where have you been? Where are you going to? J Med Chem. 2014;57:4977–5010. https://doi.org/10.1021/jm4004285.
Toropova AP, Toropov AA, Rasulev BF, Benfenati E, Gini G, Leszczynska D, et al. QSAR models for ACE-inhibitor activity of tri-peptides based on representation of the molecular structure by graph of atomic orbitals and SMILES. Struct Chem. 2012;23:1873–8. https://doi.org/10.1007/s11224-012-9996-z.
Toropov AA, Toropova AP, Raska I, Benfenati E, Gini G. QSAR modeling of endpoints for peptides which is based on representation of the molecular structure by a sequence of amino acids. Struct Chem. 2012;23:1891–904. https://doi.org/10.1007/s11224-012-9995-0.
Toropova MA, Veselinović AM, Veselinović JB, Stojanović DB, Toropov AA. QSAR modeling of the antimicrobial activity of peptides as a mathematical function of a sequence of amino acids. Comput Biol Chem. 2015;59:126–30. https://doi.org/10.1016/j.compbiolchem.2015.09.009.
Nongonierma AB, FitzGerald RJ. Strategies for the discovery and identification of food protein-derived biologically active peptides. Trends Food Sci Technol. 2017;69:289–305. https://doi.org/10.1016/j.tifs.2017.03.003.
Nongonierma AB, FitzGerald RJ. Learnings from quantitative structure–activity relationship (QSAR) studies with respect to food protein-derived bioactive peptides: a review. RSC Adv. 2016;6:75400–13. https://doi.org/10.1039/C6RA12738J.
Zhang J, Xin L, Shan B, Chen W, Xie M, Yuen D, et al. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol Cell Proteomics. 2012;11:M111.010587. https://doi.org/10.1074/mcp.M111.010587.
Nongonierma AB, Paolella S, Mudgil P, Maqsood S, FitzGerald RJ. Identification of novel dipeptidyl peptidase IV (DPP-IV) inhibitory peptides in camel milk protein hydrolysates. Food Chem. 2018;244:340–8. https://doi.org/10.1016/j.foodchem.2017.10.033.
Vukic VR, Vukic DV, Milanovic SD, Ilicic MD, Kanuric KG, Johnson MS. In silico identification of milk antihypertensive di- and tripeptides involved in angiotensin I–converting enzyme inhibitory activity. Nutr Res. 2017;46:22–30. https://doi.org/10.1016/j.nutres.2017.07.009.
García-Moreno PJ, Espejo-Carpio FJ, Guadix A, Guadix EM. Production and identification of angiotensin I-converting enzyme (ACE) inhibitory peptides from Mediterranean fish discards. J Funct Foods. 2015;18:95–105. https://doi.org/10.1016/j.jff.2015.06.062.
Majumder K, Wu J. A new approach for identification of novel antihypertensive peptides from egg proteins by QSAR and bioinformatics. Food Res Int. 2010;43:1371–8. https://doi.org/10.1016/j.foodres.2010.04.027.
Mooney C, Haslam NJ, Pollastri G, Shields DC (2012) Towards the improved discovery and design of functional peptides: common features of diverse classes permit generalized prediction of bioactivity. PLoS One. https://doi.org/10.1371/journal.pone.0045012.
Waghu FH, Gopi L, Barai RS, Ramteke P, Nizami B, Idicula-Thomas S (2014) CAMP: collection of sequences and structures of antimicrobial peptides. Nucleic Acids Res. https://doi.org/10.1093/nar/gkt1157.
Waghu FH, Barai RS, Gurung P, Idicula-Thomas S. CAMPR3: a database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2016;44:D1094–7. https://doi.org/10.1093/nar/gkv1051.
O’Keeffe MB, Fitzgerald RJ. Identification of short peptide sequences in complex milk protein hydrolysates. Food Chem. 2015;184:140–6. https://doi.org/10.1016/j.foodchem.2015.03.077.
Yesmine BH, Antoine B, da Silva Ortência Leocádia NG, Rogério BW, Ingrid A, Nicolas B, et al. Identification of ace inhibitory cryptides in Tilapia protein hydrolysate by UPLC–MS/MS coupled to database analysis. J Chromatogr B Anal Technol Biomed Life Sci. 2017;1052:43–50. https://doi.org/10.1016/j.jchromb.2017.02.015.
Lafarga T, Hayes M. Bioactive protein hydrolysates in the functional food ingredient industry: overcoming current challenges. Food Rev Int. 2017;33:217–46. https://doi.org/10.1080/87559129.2016.1175013.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Published in the topical collection Discovery of Bioactive Compounds with guest editors Aldo Laganà, Anna Laura Capriotti and Chiara Cavaliere.
Rights and permissions
About this article
Cite this article
Piovesana, S., Capriotti, A.L., Cavaliere, C. et al. Recent trends and analytical challenges in plant bioactive peptide separation, identification and validation . Anal Bioanal Chem 410, 3425–3444 (2018). https://doi.org/10.1007/s00216-018-0852-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-018-0852-x