Abstract
Aims/hypothesis
Exposure of pancreatic beta cells to cytokines released by islet-infiltrating immune cells induces alterations in gene expression, leading to impaired insulin secretion and apoptosis in the initial phases of type 1 diabetes. Long non-coding RNAs (lncRNAs) are a new class of transcripts participating in the development of many diseases. As little is known about their role in insulin-secreting cells, this study aimed to evaluate their contribution to beta cell dysfunction.
Methods
The expression of lncRNAs was determined by microarray in the MIN6 beta cell line exposed to proinflammatory cytokines. The changes induced by cytokines were further assessed by real-time PCR in islets of control and NOD mice. The involvement of selected lncRNAs modified by cytokines was assessed after their overexpression in MIN6 cells and primary islet cells.
Results
MIN6 cells were found to express a large number of lncRNAs, many of which were modified by cytokine treatment. The changes in the level of selected lncRNAs were confirmed in mouse islets and an increase in these lncRNAs was also seen in prediabetic NOD mice. Overexpression of these lncRNAs in MIN6 and mouse islet cells, either alone or in combination with cytokines, favoured beta cell apoptosis without affecting insulin production or secretion. Furthermore, overexpression of lncRNA-1 promoted nuclear translocation of nuclear factor of κ light polypeptide gene enhancer in B cells 1 (NF-κB).
Conclusions/interpretation
Our study shows that lncRNAs are modulated during the development of type 1 diabetes in NOD mice, and that their overexpression sensitises beta cells to apoptosis, probably contributing to their failure during the initial phases of the disease.
Similar content being viewed by others
Introduction
Fine-tuning of insulin release from beta cells is essential to maintain blood glucose homeostasis. Type 1 diabetes is an autoimmune disease characterised by progressive infiltration of pancreatic islets by mononuclear cells, an inflammatory process called insulitis which leads to gradual destruction of beta cells [1]. During the initial phases of the disease, beta cells are chronically exposed to cytokines and other pro-apoptotic mediators, released by the immune cells infiltrating the islets of Langerhans and by the islet cells themselves [1]. Prolonged exposure to proinflammatory cytokines such as IL-1β, TNF-α and IFN-γ has deleterious effects on specialised beta cell functions, which leads to a decreased capacity to produce and release insulin in response to secretagogues, and ultimately to beta cell loss by apoptosis [1–4]. A deeper understanding of the mechanisms occurring during this inflammatory process is of paramount importance to identify new strategies for preventing and treating the disease.
Chronic exposure of beta cells to cytokines is known to modulate the expression of many genes, resulting in severe impairment of key signalling pathways [5–7]. Previous studies focused mainly on protein-coding genes, giving minor attention to a newly identified set of regulatory factors, the non-coding RNAs. In the context of diabetes, the most studied non-coding RNAs are the microRNAs (miRNAs) [8]. These molecules, which are important controllers of gene networks, are critical regulators of specialised beta cell functions [8] and are involved in beta cell damage during the development of type 1 diabetes [9, 10]. In addition to miRNAs, recent transcriptome analysis has identified another class of functional molecules, the long non-coding RNAs (lncRNAs) [11–13]. Although the role of lncRNAs remains largely unknown, members of this RNA family have been involved in diverse gene-regulatory mechanisms such as transcription, imprinting, splicing, protein degradation and epigenetic marks on chromatin [14–19], and their dysregulation has been implicated in many human diseases [20, 21]. Little is known about the role of lncRNAs in the maintenance of beta cell activities and there are no data regarding the possible contribution of these molecules to the development of type 1 diabetes.
The aim of this project was to identify the lncRNAs that are differentially expressed under pathophysiological conditions favouring the development of type 1 diabetes, and to study their possible involvement in cytokine-mediated beta cell damage. We found that proinflammatory cytokines induce considerable changes in the transcriptional landscape of lncRNAs, some of which were also confirmed in islets of prediabetic NOD mice, a well-known model of type 1 diabetes [22]. Studies on the beta cell line MIN6 and in dissociated islet cells showed that overexpression of a selected subset of lncRNAs, either alone or in combination with cytokines, promotes beta cell apoptosis. In this study, we show for the first time that lncRNAs are modulated by proinflammatory cytokines and contribute to beta cell demise during the initial phases of murine type 1 diabetes.
Methods
Chemicals
Recombinant mouse IL-1β, hexadimethrine bromide, collagenase and Histopaque 1119 and 1077 were purchased from Sigma-Aldrich (St Louis, MO, USA). Recombinant mouse TNF-α was purchased from Enzo Life Sciences (Farmingdale, NY, USA), and recombinant mouse IFN-γ from R&D Systems (Minneapolis, MN, USA). Hoechst dye 33342 was from Invitrogen (Basel, Switzerland).
Isolation, culture and dissociation of primary islet cells
Mouse pancreatic islets were isolated from female NOD mice (Jackson Laboratory, Bar Harbor, ME, USA), male NOD severe combined immunodeficiency (SCID) mice (Janvier Labs, Le Genest-Saint-Isle, France) and C57BL/6 mice (13–14 weeks old; Charles River Laboratories, L’Arbresle, France). All animal procedures were performed in accordance with the National Institutes of Health guidelines and protocols were approved by the Swiss research council and veterinary offices. Mice islets were isolated by collagenase digestion [23] of the pancreas followed by Histopaque density gradient separation. Details about islet handling are provided in electronic supplementary material (ESM) Methods.
MIN6B1 cell culture
The murine insulin-secreting cell line MIN6B1 [24] was cultured in DMEM-Glutamax medium (Invitrogen), supplemented with 15% (vol./vol.) FCS, 50 μg/ml streptomycin, 50 U/ml penicillin (Invitrogen) and 70 μmol/l β-mercaptoethanol (Sigma). Cells were seeded at a density of 105/cm2 for insulin secretion and RNA isolation, and at 5 × 104/cm2 for cell death measurements.
Microarray profiling
Total RNA was isolated with the RNeasy kit (Qiagen, Basel, Switzerland) from MIN6 cells treated with 0.1 ng/ml IL-1β, 10 ng/ml TNF-α and 30 ng/ml IFN-γ for 24 h. Global mRNA and lncRNA expression profiling using the Mouse LncRNA Array v2.0 and data analysis were carried out by Arraystar (Rockville, MD, USA). Differentially expressed lncRNAs and mRNAs were identified through Volcano Plot filtering (fold change ≥1.5, p ≤ 0.05).
Measurement of lncRNA expression
Total RNA was isolated using the RNeasy kit followed by DNase treatment on column (Qiagen). RNA, 1 μg, was reverse transcribed using Moloney murine leukaemia virus (M-MLV) reverse transcriptase, RNAse H minus (Promega, Madison, WI, USA). Real-timex PCR was performed using iQ SYBR Green mix and samples were amplified using the CFX Connect Real-Time PCR System (Biorad, Hercules, CA, USA). The full primer list is presented in ESM Table 1.
Transfection and lentiviral infection
MIN6B1 cells were transfected with a control pcDNA3 plasmid or with the same plasmid containing the sequences to translate the lncRNAs (pcDNA3_lncRNA-1; pcDNA3_lncRNA-2, pcDNA3_lncRNA-3, pcDNA3_lncRNA-4), using Lipofectamine 2000 (Invitrogen). Plasmids containing the sequence of the lncRNA of interest or a gfp mRNA were used to produce lentiviral vectors, as described [25]. Details of lentiviral manipulation are provided in ESM Methods.
Insulin secretion
Insulin secretion from MIN6B1 cells transfected with the relevant plasmids for 48 h was carried out as previously described [10].
Assessment of cell death
The percentage of apoptotic cells was determined by scoring the fraction of cells displaying pycnotic nuclei under fluorescence microscopy (AxioCam MRc5, Zeiss, Feldbach, Switzerland) after incubation with 1 μg/ml Hoechst dye. At least 500 cells were inspected for each condition.
NF-κB nuclear translocation
MIN6B1 cells were transfected with a plasmid expressing a green fluorescent protein (GFP)-tagged form of the human nuclear factor of κ light polypeptide gene enhancer in B cells 1 (NF-κB) subunit p65 (Rela) and/or the plasmid expressing the lncRNA of choice. At 24 h after transfection, the cells were treated with the indicated cytokines for 3 h, fixed and mounted on a cover slip for microscopic examination. The fraction of cells displaying nuclear localisation of p65 was calculated after analysing around 1,500 cells for each condition.
Statistical analysis
Data are presented as mean ± SEM. Statistical differences were assessed by two-tailed paired Student’s t test when only two sets of data were present (Figs 2 and 6a–d) or by one-way ANOVA followed by Tukey’s post hoc test, with a discriminating p value of 0.05 (GraphPad Prism 5, San Diego, California, USA).
Results
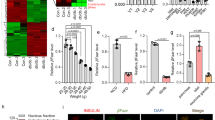
To investigate the possible contribution of lncRNAs to beta cell dysfunction, MIN6 cells were incubated for 24 h with a mix of cytokines. The expression profile of protein-coding genes and of previously annotated lncRNAs was determined by microarray analysis. In agreement with previous reports in INS1E cells [5, 26], rat beta cells [6], mice islets [27] and human islets [7], cytokines modified the expression of numerous protein-coding genes involved in inflammatory responses (i.e. Ccl2, Cxcl1, Cxcl2, Icam1, IL15), IFN-γ signalling (i.e. Irf1, Irf7, Igtp, Stat1, Stat2, Stat3, Stat4 and Jak3), NF-κB regulation (i.e. Nfkbia, Nfkb2, Nfkbiz), endoplasmic reticulum (ER) stress and apoptosis (Atf3, Atf6, Atf2, Bid, Bik1, Casp1, Casp4, Chop [also known as Ddit3]) and others (ESM Table 2). Hierarchical clustering showed a distinguishable lncRNA expression profile in the two groups of samples obtained from MIN6 cells (Fig. 1a). Control MIN6 cells expressed 18,066 of the 31,000 transcripts included in the array. On treatment with cytokines, 467 transcripts were upregulated and 219 were downregulated (Fig. 1b and ESM Table 3). Under control conditions, the average raw expression level of lncRNAs was about three times lower than that of protein-coding genes and six times lower than that observed after exposure to cytokines (data not shown). Four upregulated lncRNAs, which for simplicity will be hereafter referred as lncRNA-1, -2, -3 and -4, were selected for further analysis. LncRNA-1, -2 and -3 were chosen because they displayed the most significant expression changes. LncRNA-4 was selected because it is intergenic and the full sequence was available. The fold changes of these lncRNAs in response to cytokine treatment, and their genomic location are displayed in Fig. 1c.
(a) Hierarchical clustering of the samples analysed by microarray. The dendrogram shows the relationship between the expression of control samples (3 replicates) and samples exposed for 24 h to 0.1 ng/ml IL-1β, 10 ng/ml TNF-α and 30 ng/ml IFN-γ (3 replicates). Red, high expression; blue, low expression. (b) Volcano plot summarising the results obtained by microarray, in which control and cytokine-exposed MIN6 cells are compared. Significantly up- or downregulated transcripts are in red. (c) Studied lncRNAs with fold changes, p values and genomic locations. Cyt Mix, cytokine mix; Ctrl, control
The increase in the four lncRNAs upregulated by cytokines was confirmed by quantitative real-time PCR (Fig. 2a–d). These lncRNAs were almost undetectable in control MIN6 cells, and after incubation with cytokines for 24 h the expression of lncRNA-1, -2, -3 and -4 increased by 1,072-, 148-, 209- and 3.8-fold, respectively (Fig. 2a–d). Similar results were observed in islets isolated from C57BL/6N mice incubated with the same cytokines for 24 h, with lncRNA-1, -2, -3 and -4 being upregulated 520-, 26-, 36- and 3.6-fold, respectively (Fig. 2e–h). Similar data were obtained when the data were normalised using 18S ribosomal RNA instead of Gapdh (ESM Fig. 1). These changes appeared to be specifically induced by cytokines, as they were not observed when MIN6 cells were incubated with palmitate or with the glucagon-like peptide-1 (GLP1) analogue exendin-4 (ESM Fig. 2). The expression of these lncRNAs increased rapidly after treatment with cytokines; the induction of lncRNA-1 and -2 was significant after 6 h treatment, whereas that of lncRNA-3 and -4 was already significant after 2 h treatment (Fig. 3). Incubation of MIN6 cells with different combinations of proinflammatory cytokines showed that IFN-γ alone was sufficient to induce the expression of lncRNA-1, lncRNA-3 and lncRNA-4 (Fig. 4). The changes seen for lncRNA-2 reached statistical significance only in the presence of a mix of cytokines (Fig. 4).
The four lncRNAs are induced by proinflammatory cytokines. MIN6 cells (a–d) and mouse islets (e–h) were incubated in the absence (0) or presence of 0.1 ng/ml IL-1β, 10 ng/ml TNF-α and 30 ng/ml IFN-γ (24 h incubation). The expression levels were measured by real-time PCR and normalised to those of Gapdh. The results are means ± SEM of three to six independent experiments. * p < 0.05, ** p < 0.01 and *** p < 0.001 vs control
Time course of the induction of the lncRNAs. MIN6 cells were incubated for the indicated periods with 0.1 ng/ml IL-1β, 10 ng/ml TNF-α and 30 ng/ml IFN-γ (a–d). The expression was measured by real-time PCR and normalised to that of Gapdh. The results are means ± SEM of three independent experiments. * p < 0.05 and *** p < 0.001 vs the control at 0 h
IFN-γ is sufficient to induce the expression of the lncRNAs. MIN6 cells were incubated in the presence or absence of different cytokines at the concentrations given in the previous figures (a–d). The expression was measured by real-time PCR and normalised to that of Gapdh. The results shown are means ± SEM of three independent experiments. ** p < 0.01 and *** p < 0.001 vs control. Ctrl, control
Subsequently, we studied whether changes in the level of islet lncRNAs precede the development of type 1 diabetes in NOD mice. Although a minor infiltration of lymphocytes has been reported as early as at 4 weeks of age, in these mice peri-insulitis (a leucocytic lesion on the pole of the islet) becomes evident between 6 and 8 weeks and by week 12 is present in the majority of islets [22, 28]. In this study, we used prediabetic female NOD mice displaying normal blood glucose levels and we isolated the islets at 4, 8 and 13 weeks of age. We found that the levels of lncRNA-1 and lncRNA-2 were significantly higher in the islets of 8 and 13 week old mice than in those of 4 week old mice. LncRNA-3 expression was also higher in the islets of 8 week old mice compared with 4 week old mice, while lncRNA-4 did not reach statistical significance (Fig. 5). The expression of these lncRNAs was not modified in NOD SCID mice that do not develop type 1 diabetes (ESM Fig. 3), demonstrating that their upregulation is linked to the autoimmune reaction.
The expression of lncRNA-1, -2 and -3 increases with insulitis in islets of NOD mice. Islets were isolated from female mice aged 4, 8 and 13 weeks. The level of the four lncRNAs was measured by real-time PCR and normalised to that of Gapdh (a–d). ** p < 0.01 vs the levels measured in 4-week-old animals. Wks, weeks
We then tested whether the changes in the expression of these lncRNAs affect specific beta cell functions. Overexpression of the four lncRNAs in MIN6 cells (ESM Fig. 4) did not modify the insulin mRNA level (ESM Fig. 5), the insulin content (Fig. 6a–d) or insulin release (Fig. 6e–h). Prolonged exposure of beta cells to cytokines sensitises them to apoptosis [1, 3]. The expression of the four lncRNAs is controlled by cytokines and rises concomitantly with the increase in insulitis (Fig. 5), suggesting that they may be involved in the events causing beta cell failure. Therefore, we investigated whether the modulation of these lncRNAs affects beta cell survival. We found that overexpression of lncRNA-3 and lncRNA-4 in MIN6 cells is sufficient to increase the number of apoptotic cells, and that this effect is not increased in the presence of cytokines (Fig. 7c, d). In contrast, overexpression of either lncRNA-1 or lncRNA-2 alone did not modify the proportion of apoptotic cells. However, the overexpression of these lncRNAs increased apoptosis when the cells were concomitantly incubated with low doses of IL-1β or TNF-α, which alone did not affect cell survival (Fig. 7a, b), suggesting that the combination of both signals is necessary to trigger apoptosis. Similar results were obtained in mice islets (Fig. 7e, f), transduced with lentiviral vectors (ESM Fig. 6). We then investigated whether the overexpression of these lncRNAs, either alone or in combination with IL-1β, modifies the level of some members of the Bcl-2 family (Bik1 and Bid) or of the downstream ER-stress-related genes Chop and Atf3. No significant effect was observed on any of these genes (ESM Fig. 7).
Overexpression of the lncRNAs does not affect insulin content or secretion. MIN6 cells were transfected for 48 h with an empty plasmid (vector) or a plasmid enabling the transcription of an individual lncRNA. The cellular insulin content was measured by ELISA (a–d). To test their secretory properties, the cells were incubated with 2 or 20 mmol/l glucose for 45 min. Basal and glucose-induced insulin release were assessed by ELISA (e–h). The results represent means ± SEM of three to six independent experiments
Overexpression of the lncRNAs triggers apoptosis. (a–d) MIN6 cells were transfected with a control plasmid (vector; grey) or with plasmids overexpressing lncRNA-1 (a); lncRNA-2 (b); lncRNA-3 (c) or lncRNA-4 (d) (black). After 24 h, the cells were incubated for another 24 h with or without single cytokines or a mix of the three at a concentration of 0.1 ng/ml IL-1β, 10 ng/ml TNF-α and 30 ng/ml IFN-γ. Dispersed mouse islet cells were transduced with a control lentiviral vector expressing GFP or a vector expressing the lncRNA of interest (e, GFP in white, lncRNA-1 in grey and lncRNA-2 in black; f, as indicated). Incubation with cytokines was performed as described above. The proportion of cells showing pycnotic nuclei was scored. Fold changes were calculated by dividing the values by the percentage of pycnotic nuclei observed in controls (a–d). The results are the means±SEM of four to six independent experiments. * p < 0.05 and ** p < 0.01 vs control. Ctrl, control; Cyt Mix, cytokine mix
Although basal NF-κB activity is required for normal insulin release [29], induction of the NF-κB pathway plays a central role in cytokine-induced apoptosis [30]. Translocation of NF-κB to the nucleus is one of the initial events occurring shortly after exposure to IL-1β [31]. In view of the potential crosstalk with the NF-κB pathway, we transfected MIN6 cells with a GFP-tagged form of p65, one of the NF-κB subunits in the predominant dimers (p65/p65 and p65/p50) found in IL-1β-exposed beta cells [32], and we monitored its translocation to the nucleus on overexpression of each of the four lncRNAs. We found that incubation of cells in the presence of low doses of IL-1β (0.1 ng/ml) or IFN-γ (30 ng/ml) does not affect the intracellular distribution of NF-κB (Fig. 8, ESM Fig. 8). In contrast, incubation with higher doses of IL-1β (10 ng/ml) or IFN-γ (300 ng/ml) led to an increase in the fraction of cells in which NF-κB is localised in the nucleus. Of the four lncRNAs studied, only overexpression of lncRNA-1 mimicked the effect of high doses of IL-1β or IFN-γ, and increased the fraction of cells in which p65 is localised to the nucleus (Fig. 8). This effect was not further potentiated by exposing the cells to IL-1β (Fig. 8).
Overexpression of lncRNA-1 induces the nuclear translocation of NF-κB. MIN6 cells were transfected for 24 h with a plasmid expressing GFP-tagged p65 (Rela). They were then exposed for 3 h to the indicated cytokines before fixation and fluorescence microscopy analysis. The number of cells displaying nuclear NF-κB localisation were scored. Data were normalised to the values of control p65 with no cytokine exposure. The results shown represent means ± SEM of six independent experiments. * p < 0.05 vs the control value (p65, no cytokine exposure). Ctrl, control
Discussion
Type 1 diabetes is characterised by progressive islet infiltration by immune cells releasing cytokines and other pro-apoptotic mediators that contribute to beta cell death during the initial phases of the disease [1]. In diabetes-prone NOD mice beta cell apoptosis precedes substantial lymphocytic infiltration [33, 34], supporting the notion that inflammatory mediators released by infiltrating cells play a major role in beta cell death. Accordingly, chronic exposure to proinflammatory cytokines has detrimental effects on specialised beta cell functions [1–4] and affects the expression of protein-coding genes and of small non-coding RNAs [5–7]. We previously showed that changes in the level of miRNAs can affect the secretory capacities of beta cells and their sensitivity to apoptosis [9, 10]. Here, we not only demonstrate that incubation of MIN6 cells with a mix of T helper 1 cytokines results in considerable changes in the expression of protein-coding genes involved in inflammatory responses, IFN-γ signalling, NF-κB regulation, ER stress and apoptosis similar to those shown in previous reports [5–7, 26, 27] but we also reveal for the first time that it causes major modifications in lncRNA expression. Despite extensive research, the role of the majority of these transcripts is still unknown and no information is available regarding their contribution to the maintenance of beta cell function and the development of type 1 diabetes. We found that MIN6 cells express many previously annotated lncRNAs, and that a considerable fraction of these transcripts are modulated in the presence of proinflammatory cytokines. In agreement with previous studies [35, 36], we found that the average levels of lncRNA expression in MIN6 cells were lower than those of protein-coding genes.
We focused on four lncRNAs and demonstrated that the proinflammatory cytokines modulate their expression in mouse islets. IFN-γ was sufficient to induce the expression of these lncRNAs. Interestingly, in NOD mice during the initial phases of the disease, the islet genes displaying the higher expression changes are all induced by IFNs [28]. The expression of the four tested lncRNAs is rapidly increased by cytokines, suggesting that these lncRNAs act early in the cascade of events activated by the inflammatory mediators, and culminating in cell death. This is not surprising as the expression of many protein-coding genes involved in beta cell apoptosis is already modified after 2 h exposure to cytokine [5, 26].
Subsequently, we investigated whether changes in the level of these lncRNAs precede the development of diabetes and we measured their expression in islets of normoglycaemic NOD mice of increasing age. We found that the levels of lncRNA-1, lncRNA-2 and lncRNA-3 increase with age, paralleling the development of insulitis in the early phases of the disease [22, 28]. This was not observed in NOD SCID mice, which are immunodeficient and do not develop diabetes, suggesting that the induction of these lncRNAs is a consequence of the autoimmune reaction. In NOD mice autoimmune destruction of beta cells is preceded by defective insulin secretion [37]. However, none of the lncRNAs investigated had an impact on insulin biosynthesis and release. As in NOD mice beta cell apoptosis precedes substantial lymphocytic infiltration [33, 34] and prolonged exposure to cytokines sensitises beta cells to apoptosis [1–4], we assessed whether the modulation of lncRNAs affects the survival of insulin-secreting cells. Indeed, overexpression of lncRNA-3 and lncRNA-4 was sufficient to increase apoptosis of MIN6 and mouse islet cells. In contrast, lncRNA-1 and lncRNA-2 induced apoptosis only in cells concomitantly exposed to low doses of IL-1β or TNF-α. This suggests that lncRNA-1 and -2 trigger apoptosis only when the beta cells receive a combination of signals generated by the activation of different inflammatory pathways. This is reminiscent of the events occurring in the initial phases of type 1 diabetes when beta cell death is believed to be elicited by prolonged exposure to a combination of inflammatory mediators, rather than to low doses of a single cytokine [38]. Cytokines modify the level of many genes belonging to the Bcl2 family [6, 39] and activate the ER stress response [40]. Overexpression of the four lncRNAs, alone or in combination with IL1-β, neither altered the expression of the pro-apoptotic Bcl2 family members Bik1 and Bid, nor affected the levels of the downstream ER stress-related genes Chop and Atf3, suggesting that they may not operate through activation of these genes.
Stimulation of beta cells with either IL-1β or TNF-α results in the activation of NF-κB [41], a key regulator of gene networks controlling cytokine-induced beta cell dysfunction and death [30]. Indeed, inhibition of NF-κB activation in vitro by overexpression of inhibitory κB (IκB) protects beta cells against cytokine-induced apoptosis [42, 43]. Furthermore, beta cell-specific NF-kB blockade in vivo prevents streptozotocin-induced diabetes development [44]. The translocation of NF-κB into the nucleus is one of the initial events occurring shortly after exposure to IL-1β or TNF-α [31]. In INS1E cells, IL-1β triggers a stronger induction of NF-κB target genes compared with TNF-α, partly because it elicits an earlier translocation of NF-κB into the nucleus [31] and a more stable activation of this signalling pathway [32]. We found that lncRNA-1 promotes the translocation of p65 into the nucleus, mimicking the effects observed when the cells are exposed to high doses of IL-1β and IFN-γ. On overexpression of lncRNA-1, low doses of IL-1β are sufficient to induce apoptosis. Thus, sensitisation of beta cells overexpressing lncRNA-1 to apoptosis may at least in part be caused by NF-κB nuclear translocation. Our findings indicate a possible crosstalk of IFN-γ with the NF-κB pathway in beta cells. IFN-γ exerts its functions mainly through the Janus kinase (JAK)-signal transducer and activator of transcription (JAK-STAT) cascade [45], but accumulating evidence indicates that in some cell types IFN-γ can also activate the NF-κB pathway [46, 47]. The mechanisms through which the NF-κB pathway may mediate the actions of IFN-γ remain largely unknown. Our data suggest that lncRNAs could provide a link between the two signalling cascades. Further studies will be needed to elucidate the precise mechanism through which lncRNA-1 induces the nuclear translocation of NF-κB and, in general, to understand the role of lncRNA-2, lncRNA-3 and lncRNA-4 in beta cell apoptosis.
Overall, our findings show that proinflammatory cytokines induce extensive changes in the expression of previously annotated lncRNAs. The study of four selected lncRNAs shows that the expression of these non-coding transcripts increases with the development of insulitis in NOD mice and that their modulation increases beta cell apoptosis, suggesting that they may contribute to cytokine-mediated beta cell dysfunction occurring during the initial phases of type 1 diabetes. The four lncRNAs investigated in this study are most probably not the only long non-coding transcripts contributing to beta cell demise. In fact, our data have highlighted dramatic changes in the level of many other lncRNAs elicited by cytokines and these inflammatory mediators may potentially also affect the expression of some of the recently described beta cell-specific mouse lncRNAs [48] that were not included in the array.
Future studies will not only need to elucidate the contribution of all these lncRNAs in the development of type 1 diabetes in mice but will also need to assess whether the same mechanism operates in humans. This promises to be a difficult task because, although the function of lncRNAs is likely to be maintained, the conservation of the sequence throughout species can be as little as 21%, with exons being under less evolutionary constraint than in protein-coding genes [35, 49]. The genomic loci from which the four lncRNAs are generated produce a large number of partially overlapping transcripts and it was not possible to identify with certainty their human orthologues. New computational approaches based on the analysis of the secondary structure rather than on sequence conservation will be necessary to precisely identify the human transcripts corresponding to the lncRNAs investigated in this study.
Abbreviations
- ER:
-
Endoplasmic reticulum
- GFP:
-
Green flourescent protein
- lncRNA:
-
Long non-coding RNA
- miRNA:
-
MicroRNA
- NF-κB:
-
Nuclear factor of κ light polypeptide gene enhancer in B cells 1
- Rela:
-
NF-κB subunit p65
- SCID:
-
Severe combined immunodeficiency
References
Eizirik DL, Colli ML, Ortis F (2009) The role of inflammation in insulitis and beta-cell loss in type 1 diabetes. Nat Rev Endocrinol 5:219–226
Donath MY, Storling J, Berchtold LA, Billestrup N, Mandrup-Poulsen T (2008) Cytokines and beta-cell biology: from concept to clinical translation. Endocr Rev 29:334–350
Pipeleers D, Hoorens A, Marichal-Pipeleers M, van de Casteele M, Bouwens L, Ling Z (2001) Role of pancreatic beta-cells in the process of beta-cell death. Diabetes 50(Suppl 1):S52–S57
Kaminitz A, Stein J, Yaniv I, Askenasy N (2007) The vicious cycle of apoptotic beta-cell death in type 1 diabetes. Immunol Cell Biol 85:582–589
Kutlu B, Cardozo AK, Darville MI et al (2003) Discovery of gene networks regulating cytokine-induced dysfunction and apoptosis in insulin-producing INS-1 cells. Diabetes 52:2701–2719
Ortis F, Naamane N, Flamez D et al (2010) Cytokines interleukin-1beta and tumor necrosis factor-alpha regulate different transcriptional and alternative splicing networks in primary beta-cells. Diabetes 59:358–374
Eizirik DL, Sammeth M, Bouckenooghe T et al (2012) The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLoS Genet 8:e1002552
Guay C, Jacovetti C, Nesca V, Motterle A, Tugay K, Regazzi R (2012) Emerging roles of non-coding RNAs in pancreatic beta-cell function and dysfunction. Diabetes Obes Metab 14(Suppl 3):12–21
Roggli E, Britan A, Gattesco S et al (2010) Involvement of microRNAs in the cytotoxic effects exerted by proinflammatory cytokines on pancreatic beta-cells. Diabetes 59:978–986
Roggli E, Gattesco S, Caille D et al (2012) Changes in microRNA expression contribute to pancreatic beta-cell dysfunction in prediabetic NOD mice. Diabetes 61:1742–1751
Carninci P, Kasukawa T, Katayama S et al (2005) The transcriptional landscape of the mammalian genome. Science 309:1559–1563
Mattick JS (2009) The genetic signatures of noncoding RNAs. PLoS Genet 5:e1000459
Consortium EP (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74
Wang KC, Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43:904–914
Guttman M, Donaghey J, Carey BW et al (2011) lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477:295–300
Khaitan D, Dinger ME, Mazar J et al (2011) The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res 71:3852–3862
Ginger MR, Shore AN, Contreras A et al (2006) A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc Natl Acad Sci U S A 103:5781–5786
Paralkar VR, Weiss MJ (2011) A new 'Linc' between noncoding RNAs and blood development. Genes Dev 25:2555–2558
Tripathi V, Ellis JD, Shen Z et al (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39:925–938
Wapinski O, Chang HY (2011) Long noncoding RNAs and human disease. Trends Cell Biol 21:354–361
Batista PJ, Chang HY (2013) Long noncoding RNAs: cellular address codes in development and disease. Cell 152:1298–1307
Debussche X, Lormeau B, Boitard C, Toublanc M, Assan R (1994) Course of pancreatic beta cell destruction in prediabetic NOD mice: a histomorphometric evaluation. Diabetes Metab 20:282–290
Gotoh M, Maki T, Satomi S et al (1987) Reproducible high yield of rat islets by stationary in vitro digestion following pancreatic ductal or portal venous collagenase injection. Transplantation 43:725–730
Lilla V, Webb G, Rickenbach K et al (2003) Differential gene expression in well-regulated and dysregulated pancreatic beta-cell (MIN6) sublines. Endocrinology 144:1368–1379
Marr RA, Rockenstein E, Mukherjee A et al (2003) Neprilysin gene transfer reduces human amyloid pathology in transgenic mice. J Neurosci 23:1992–1996
Moore F, Naamane N, Colli ML et al (2011) STAT1 is a master regulator of pancreatic β-cell apoptosis and islet inflammation. J Biol Chem 286:929–941
Wolden-Kirk H, Rondas D, Bugliani M et al (2014) Discovery of molecular pathways mediating 1,25-dihydroxyvitamin D3 protection against cytokine-induced inflammation and damage of human and male mouse islets of Langerhans. Endocrinology 155:736–747
Carrero JA, Calderon B, Towfic F, Artyomov MN, Unanue ER (2013) Defining the transcriptional and cellular landscape of type 1 diabetes in the NOD mouse. PLoS One 8:e59701
Norlin S, Ahlgren U, Edlund H (2005) Nuclear factor-{kappa}B activity in β-cells is required for glucose-stimulated insulin secretion. Diabetes 54:125–132
Cardozo AK, Heimberg H, Heremans Y et al (2001) A comprehensive analysis of cytokine-induced and nuclear factor-kappa B-dependent genes in primary rat pancreatic beta cells. J Biol Chem 276:48879–48886
Ortis F, Pirot P, Naamane N et al (2008) Induction of nuclear factor-kappaB and its downstream genes by TNF-alpha and IL-1beta has a pro-apoptotic role in pancreatic beta cells. Diabetologia 51:1213–1225
Ortis F, Cardozo AK, Crispim D, Storling J, Mandrup-Poulsen T, Eizirik DL (2006) Cytokine-induced proapoptotic gene expression in insulin-producing cells is related to rapid, sustained, and nonoscillatory nuclear factor-kappaB activation. Mol Endocrinol 20:1867–1879
Kurrer MO, Pakala SV, Hanson HL, Katz JD (1997) Beta cell apoptosis in T cell-mediated autoimmune diabetes. Proc Natl Acad Sci U S A 94:213–218
O'Brien BA, Harmon BV, Cameron DP, Allan DJ (1997) Apoptosis is the mode of beta-cell death responsible for the development of IDDM in the nonobese diabetic (NOD) mouse. Diabetes 46:750–757
Derrien T, Johnson R, Bussotti G et al (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22:1775–1789
Ravasi T, Suzuki H, Pang KC et al (2006) Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res 16:11–19
Ize-Ludlow D, Lightfoot YL, Parker M et al (2011) Progressive erosion of beta-cell function precedes the onset of hyperglycemia in the NOD mouse model of type 1 diabetes. Diabetes 60:2086–2091
Eizirik DL, Mandrup-Poulsen T (2001) A choice of death—the signal-transduction of immune-mediated beta-cell apoptosis. Diabetologia 44:2115–2133
Gurzov EN, Eizirik DL (2011) Bcl-2 proteins in diabetes: mitochondrial pathways of beta-cell death and dysfunction. Trends Cell Biol 21:424–431
Eizirik DL, Cardozo AK, Cnop M (2008) The role for endoplasmic reticulum stress in diabetes mellitus. Endocr Rev 29:42–61
Hayden MS, Ghosh S (2004) Signaling to NF-kappaB. Genes Dev 18:2195–2224
Giannoukakis N, Rudert WA, Trucco M, Robbins PD (2000) Protection of human islets from the effects of interleukin-1beta by adenoviral gene transfer of an Ikappa B repressor. J Biol Chem 275:36509–36513
Heimberg H, Heremans Y, Jobin C et al (2001) Inhibition of cytokine-induced NF-kappaB activation by adenovirus-mediated expression of a NF-kappaB super-repressor prevents beta-cell apoptosis. Diabetes 50:2219–2224
Eldor R, Yeffet A, Baum K et al (2006) Conditional and specific NF-kappaB blockade protects pancreatic beta cells from diabetogenic agents. Proc Natl Acad Sci U S A 103:5072–5077
Gough DJ, Levy DE, Johnstone RW, Clarke CJ (2008) IFNgamma signaling-does it mean JAK-STAT? Cytokine Growth Factor Rev 19:383–394
Lin Y, Jamison S, Lin W (2012) Interferon-gamma activates nuclear factor-kappa B in oligodendrocytes through a process mediated by the unfolded protein response. PLoS One 7:e36408
Rimbach G, Valacchi G, Canali R, Virgili F (2000) Macrophages stimulated with IFN-gamma activate NF-kappa B and induce MCP-1 gene expression in primary human endothelial cells. Mol Cell Biol Res Commun 3:238–242
Ku GM, Kim H, Vaughn IW et al (2012) Research resource: RNA-Seq reveals unique features of the pancreatic beta-cell transcriptome. Mol Endocrinol 26:1783–1792
Cabili MN, Trapnell C, Goff L et al (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25:1915–1927
Acknowledgements
We would like to thank the group of C. Widmann for kindly providing the vector expressing Rela and Arraystar for the microarray service provided.
Funding
This work was supported by the European Foundation for the Study of Diabetes and Lilly research fellowship (AM), by grants of the Swiss National Science Foundation (310030-146138 to RR and 310000-141162 to PM) and by the Fondation Francophone pour la Recherche sur le Diabète (RR).
Duality of interest
The authors declare that there is no duality of interest associated to this manuscript.
Contribution statement
AM conceived the experiments, generated the research, analysed the data, wrote the manuscript and approved its final version. SG, DC and PM contributed to the acquisition of data, reviewed the manuscript and approved its final version. RR conceived the experiments, analysed the research data, wrote the manuscript and approved its final version. AM is responsible for the integrity of the work as a whole.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM Methods
(PDF 113 kb)
ESM Table 1
(PDF 34 kb)
ESM Table 2
(PDF 84 kb)
ESM Table 3
(PDF 88 kb)
ESM Fig. 1
(PDF 86.1 kb)
ESM Fig. 2
(PDF 69.3 kb)
ESM Fig. 3
(PDF 68.4 kb)
ESM Fig. 4
(PDF 69.5 kb)
ESM Fig. 5
(PDF 67.1 kb)
ESM Fig. 6
(PDF 69.5 kb)
ESM Fig. 7
(PDF 119 kb)
ESM Fig. 8
(PDF 171 kb)
Rights and permissions
About this article
Cite this article
Motterle, A., Gattesco, S., Caille, D. et al. Involvement of long non-coding RNAs in beta cell failure at the onset of type 1 diabetes in NOD mice. Diabetologia 58, 1827–1835 (2015). https://doi.org/10.1007/s00125-015-3641-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-015-3641-5