Abstract
Key message
A novel locus, qCSS3, involved in the non-seed-shattering behaviour of Japonica rice cultivar, ‘Nipponbare’, was detected by QTL-seq analysis using the segregating population with the fixed known seed-shattering loci.
Abstract
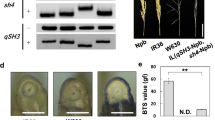
Asian cultivated rice, Oryzasativa, was domesticated from its wild ancestor, O.rufipogon. Loss of seed shattering is one of the most recognisable traits selected during rice domestication. Three quantitative trait loci (QTLs), qSH1, qSH3, and sh4, were previously reported to be involved in the loss of seed shattering of Japonica cultivated rice, O.sativa ‘Nipponbare’. However, the introgression line (IL) carrying ‘Nipponbare’ alleles at these three loci in the genetic background of wild rice, O.rufipogon W630, showed a lower value for detaching a grain from the pedicel than ‘Nipponbare’. Here, we investigated abscission layer formation in the IL and found a partially formed abscission layer in the central region between the epidermis and vascular bundles. Based on QTL-seq analysis using the F2 population obtained from a cross between ‘Nipponbare’ and the IL, we detected two novel loci qCSS3 and qCSS9 (QTL for the Control of Seed Shattering in rice on chromosomes 3 and 9), which were found to be involved in the difference in seed-shattering degree between ‘Nipponbare’ and W630. Then, we further focused on qCSS3 in order to understand its potential role on the loss of seed shattering. The candidate region of qCSS3 was found to be located within a 526-kb region using substitution mapping analysis. Interestingly, the qCSS3 candidate region partially overlaps the selective sweep detected for Japonica but not for Indica rice cultivars, suggesting that this region harbours the mutation at a novel seed-shattering locus specifically selected for non-seed-shattering behaviour in Japonica cultivars.




Similar content being viewed by others
References
Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, Innan H, Cano L, Kamoun S, Terauchi R (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30:174–178. https://doi.org/10.1038/nbt.2095
Doebley JF, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication. Cell 127:1309–1321. https://doi.org/10.1016/j.cell.2006.12.006
Dong Y, Wang YZ (2015) Seed shattering: from models to crops. Front Plant Sci 6:476. https://doi.org/10.3389/fpls.2015.00476
Fuller DQ (2007) Contrasting patterns in crop domestication and domestication rates: recent archaeobotanical insights from the Old World. Ann Bot 100:903–924. https://doi.org/10.1093/aob/mcm048
Fuller DQ, Allaby R (2009) Seed dispersal and crop domestication: shattering, germination and seasonality in evolution under cultivation. In: Østergaard L (ed) Annual plant reviews, vol 38: fruit development and seed dispersal. Wiley, Oxford, pp 238–295
Harlan JR (1975) Crops and man. American Society of Agronomy, Madison
Htun TM, Inoue C, Chhourn O, Ishii T, Ishikawa R (2014) Effect of quantitative trait loci for seed shattering on abscission layer formation in Asian wild rice Oryza rufipogon. Breed Sci 64:199–205. https://doi.org/10.1270/jsbbs.64.199
Huang X, Kurata N, Wei X, Wang ZX, Wang A, Zhao Q, Zhao Y, Liu K, Lu H, Li W, Guo Y, Lu Y, Zhou C, Fan D, Weng Q, Zhu C, Huang T, Zhang L, Wang Y, Feng L, Furuumi H, Kubo T, Miyabayashi T, Yuan X, Xu Q, Dong G, Zhan Q, Li C, Fujiyama A, Toyoda A, Lu T, Feng Q, Qian Q, Li J, Han B (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature 490:497–501. https://doi.org/10.1038/nature11532
Inoue C, Htun TM, Inoue K, Ikeda K, Ishii T, Ishikawa R (2015) Inhibition of abscission layer formation by an interaction of two seed-shattering loci, sh4 and qSH3, in rice. Genes Genet Syst 90:1–9. https://doi.org/10.1266/ggs.90.1
Ishikawa R, Thanh PT, Nimura N, Htun TM, Yamasaki M, Ishii T (2010) Allelic interaction at seed-shattering loci in the genetic backgrounds of wild and cultivated rice species. Genes Genet Syst 85:265–271. https://doi.org/10.1266/ggs.85.265
Ishikawa R, Nishimura A, Htun TM, Nishioka R, Oka Y, Tsujimura Y, Inoue C, Ishii T (2017) Estimation of loci involved in non-shattering of seeds in early rice domestication. Genetica 145:201–207. https://doi.org/10.1007/s10709-017-9958-x
Konishi S, Izawa T, Lin SY, Ebana K, Fukuta Y, Sasaki T, Yano M (2006) An SNP caused loss of seed shattering during rice domestication. Science 312:1392–1396. https://doi.org/10.1126/science.1126410
Kosugi S, Natsume S, Yoshida K, MacLean D, Cano L, Kamoun S, Terauchi R (2013) Coval: improving alignment quality and variant calling accuracy for next-generation sequencing data. PLoS ONE 8:e75402. https://doi.org/10.1371/journal.pone.0075402
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Li C, Zhou A, Sang T (2006) Rice domestication by reducing shattering. Science 311:1936–1939. https://doi.org/10.1126/science.1123604
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Lin Z, Griffith ME, Li X, Zhu Z, Tan L, Fu Y, Zhang W, Wang X, Xie D, Sun C (2007) Origin of seed shattering in rice (Oryza sativa L.). Planta 226:11–20. https://doi.org/10.1007/s00425-006-0460-4
Oka HI (1988) Origin of cultivated rice. Elsevier, Amsterdam
Onishi K, Horiuchi Y, Ishigoh-Oka N, Takagi K, Ichikawa N, Maruoka M, Sano Y (2007) A QTL cluster for plant architecture and its ecological significance in Asian wild rice. Breed Sci 57:7–16. https://doi.org/10.1270/jsbbs.57.7
Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, Uemura A, Utsushi H, Tamiru M, Takuno S, Innan H, Cano LM, Kamoun S, Terauchi R (2013) QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 74:174–183. https://doi.org/10.1111/tpj.12105
Thanh PT, Phan PDT, Mori N, Ishikawa R, Ishii T (2011) Development of backcross recombinant inbred lines between Oryza sativa Nipponbare and O. rufipogon and QTL detection on drought tolerance. Breed Sci 61:76–79. https://doi.org/10.1270/jsbbs.61.76
Tsujimura Y, Inoue C, Htun TM, Oka Y, Ishii T, Ishikawa R (2017) Investigation of genetic loci controlling non-shattering behaviour in an Indica rice cultivar ‘IR36’. J Crop Res 62:19–23. https://doi.org/10.18964/jcr.62.0_19
Xu X, Liu X, Ge S, Jensen JD, Hu F, Li X, Dong Y, Gutenkunst RN, Fang L, Huang L, Li J, He W, Zhang G, Zheng X, Zhang F, Li Y, Yu C, Kristiansen K, Zhang X, Wang J, Wright M, McCouch S, Nielsen R, Wang J, Wang W (2012) Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat Biotechnol 30:105–111. https://doi.org/10.1038/nbt.2050
Zhang LB, Zhu Q, Wu ZQ, Ross-Ibarra J, Gaut BS, Ge S, Sang T (2009) Selection on grain shattering genes and rates of rice domestication. New Phytol 184:708–720. https://doi.org/10.1111/j.1469-8137.2009.02984.x
Acknowledgments
The wild rice accession, O. rufipogon W630, was provided by the National Institute of Genetics supported by the National Bioresource Project, MEXT, Japan. We thank Satoshi Natsume (Iwate Biotechnology Research Center), Kentaro Yoshida (Kobe University), Soichiro Nishiyama and Takuya Morimoto (Kyoto University) for their useful advices on our QTL-seq analysis. This study was supported in part by Grants-in-Aid from the Japan Society for the Promotion of Science (JSPS) to R.I. (Nos. 15KK0280 and 18K05594) and by the JSPS Bilateral Open Partnership Joint Research Project to R.I.
Author information
Authors and Affiliations
Contributions
RI conceived and designed the study. YT, SS, KO, TMT, CC, and RI performed the experiments. YT, SS, KO, TI, and RI analysed the data. YT, KN, and TA contributed to bioinformatic analysis. YT, TI, and RI prepared the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Yunbi Xu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. 1
Results of the QTL-seq analysis for all chromosomes. The Δ(SNP-index) plots with statistical intervals under the null hypothesis of no QTL (orange, P < 0.01; green; P < 0.05). The red line indicates the average ΔSNP-index calculated by a sliding window analysis (PDF 822 kb)
Supplemental Fig. 2
Box plots of the BTS values of two F2 groups based on the genotypes at the candidate regions of qCSS3 and qCSS9. All F2 individuals were surveyed with five and four PCR-based molecular markers covering qCSS3 and qCSS9 regions, respectively (Supplemental Tables 1 and 2). Plants carrying the ‘Nipponbare’ (Npb) and W630 homozygous chromosomal segments for the entire candidate regions of qCSS3 and qCSS9 were selected (PDF 327 kb)
Supplemental Fig. 3
Evaluation of seed-shattering degree of the two backcross recombinant inbred lines (BRILs) carrying the W630 chromosomal segment covering the qCSS3 region in the ‘Nipponbare’ genetic background. (a) Graphical genotypes of two BRILs, BRIL42 and BRIL86. Red lines indicate the region covering qCSS3. (b) Seed-shattering degree of ‘Nipponbare’ (Npb) and the two BRILs. Data are mean ± S.D. (n = 6). ** indicates P < 0.01 by unpaired Student’s t-test (PDF 465 kb)
Supplemental Table 1
PCR-based molecular markers used in this study (PDF 79 kb)
Supplemental Table 2
Information of each 174 F2 plant used for QTL-seq analysis (PDF 98 kb)
Supplemental Table 3
Days to heading observed for F3 progeny from three F2 plants (Nos. 57, 54, and 129). Data are mean ± S.D. (n = 6) (PDF 29 kb)
Supplemental Table 4
List of the 96 genes annotated in a 526-kb candidate region of qCSS3 (PDF 36 kb)
Rights and permissions
About this article
Cite this article
Tsujimura, Y., Sugiyama, S., Otsuka, K. et al. Detection of a novel locus involved in non-seed-shattering behaviour of Japonica rice cultivar, Oryzasativa ‘Nipponbare’. Theor Appl Genet 132, 2615–2623 (2019). https://doi.org/10.1007/s00122-019-03376-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-019-03376-3