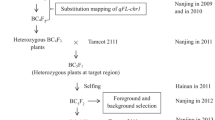
Abstract
Key message
qFS07.1 controlling fiber strength was fine-mapped to a 62.6-kb region containing four annotated genes. RT-qPCR and sequence of candidate genes identified an LRR RLK gene as the most likely candidate.
Abstract
Fiber strength is an important component of cotton fiber quality and is associated with other properties, such as fiber maturity, fineness, and length. Stable QTL qFS07.1, controlling fiber strength, had been identified on chromosome 7 in an upland cotton recombinant inbred line (RIL) population from a cross (CCRI35 × Yumian1) described in our previous studies. To fine-map qFS07.1, an F2 population with 2484 individual plants from a cross between recombinant line RIL014 and CCRI35 was established. A total of 1518 SSR primer pairs, including 1062, designed from chromosome 1 of the Gossypium raimondii genome and 456 from chromosome 1 of the G. arboreum genome (corresponding to the QTL region) were used to fine-map qFS07.1, and qFS07.1 was mapped into a 62.6-kb genome region which contained four annotated genes on chromosome A07 of G. hirsutum. RT-qPCR and comparative analysis of candidate genes revealed a leucine-rich repeat protein kinase (LRR RLK) family protein to be a promising candidate gene for qFS07.1. Fine mapping and identification of the candidate gene for qFS07.1 will play a vital role in marker-assisted selection (MAS) and the study of mechanism of cotton fiber development.






Similar content being viewed by others
References
Arpat AB, Waugh M, Sullivan JP, Gonzales M, Frisch D, Main D, Wood T, Leslie A, Wing RA, Wilkins TA (2004) Functional genomics of cell elongation in developing cotton fibers. Plant Mol Biol 54:911–929
Barrero JM, Cavanagh C, Verbyla KL, Tibbits JFG, Verbyla AP, Huang BE, Gubler F (2015) Transcriptomic analysis of wheat near-isogenic lines identifies PM19-A1 and A2 as candidates for a major dormancy QTL. Genom Biol 16(1):1
Bolon YT, Joseph B, Cannon SB, Graham MA, Diers BW, Farmer AD, May GD, Muehlbauer GJ, Specht JE, Tu ZJ, Weeks N, Xu WW, Shoemaker RC, Vance CP (2010) Complementary genetic and genomic approaches help characterize the linkage group I seed protein QTL in soybean. BMC Plant Biol 10:41
Bradow JM, Davidonis GH (2000) Quantitation of fiber quality and the cotton production-processing interface: a physiologist’s perspective. J Cotton Sci 4:34–64
Cao ZB, Zhu XF, Chen H, Zhang TZ (2015) Fine mapping of clustered quantitative trait loci for fiber quality on chromosome 7 using a Gossypium barbadense introgressed line. Mol Breed 35:215
Chen ZJ, Scheffler BE, Dennis E, Triplett BA, Zhang TZ, Guo WZ, Chen XY, Stelly DM, Rabinowicz PD, Town CD, Arioli T, Brubaker C, Cantrell RG, Lacape JM, Ulloa M, Chee P, Gingle AR, Haigler CH, Percy R, Saha S, Wilkins T, Wright RJ, Deynze AV, Zhu YX, Yu SX, Abdurakhmonov I, Katageri I, Kumar PA, Rahman M, Zafar Y, Yu JZ, Kohel RJ, Wendel JF, Paterson AH (2007) Toward sequencing cotton (Gossypium) genomes. Plant Physiol 145:1303–1310
Davis LA, Addicott FT (1972) Abscisic acid: correlation with abscission and with development in the cotton fruit. Plant Physiol 49:644–648
Deng FL, Tu LL, Tan JF, Li Y, Nie YC, Zhang XL (2012) GbPDF1 is involved in cotton fiber initiation via the core cis-element HDZIP2ATATHB2. Plant Physiol 158:890–904
Fang L, Tian RP, Chen JD, Wang S, Li XH, Wang P, Zhang TZ (2014b) Transcriptomic analysis of fiber strength in Upland cotton chromosome introgression lines carrying different Gossypium barbadense chromosomal segments. PLoS One 9:e94642
Fang DD, Jenkins JN, Deng DD, McCarty JC, Li P, Wu J (2014a) Quantitative trait loci analysis of fiber quality traits using a random-mated recombinant inbred population in Upland cotton (Gossypium hirsutum L.). BMC Genom 15:397
Gokani SJ, Kumar R, Thaker VS (1998) Potential role of abscisic acid in cotton fiber and ovule development. J Plant Growth Regul 17:1–5
Hamann T (2015) The plant cell wall integrity maintenance mechanism-a case study of a cell wall plasma membrane signaling network. Phytochem 112:100–109
Harpaz-Saad S, McFarlane HE, Xu S, Divi UK, Forward B, Western TL, Kieber JJ (2011) Cellulose synthesis via the FEI2 RLK/ SOS5 pathway and cellulose synthase 5 is required for the structure of seed coat mucilage in Arabidopsis. Plant J 68:941–953
Huang JF, Chen F, Wu SY, Li J, Xu W (2016) Cotton GhMYB7 is predominantly expressed in developing fibers and regulates secondary cell wall biosynthesis in transgenic Arabidopsis. Sci China. Life Sci 59(2):194–205
Huang GQ, Gong SY, Xu WL, Li W, Li P, Zhang CJ, Li DD, Zheng Y, Li FG, Li XB (2013) A fasciclin-like arabinogalactan protein, GhFLA1, is involved in fiber initiation and elongation of cotton. Plant Physiol 161:1278–1290
Islam MS, Zeng LH, Thyssen GN, Christoher DD, Kim HJ, Li P, Fang DD (2016) Mapping by sequencing in cotton (Gossypium hirsutum) line MD52ne identified candidate genes for fiber strength and its related quality attributes. Theor Appl Genet 129(6):1071–1086
Jiang YJ, Guo WZ, Zhu HY, Ruan YL, Zhang TZ (2012) Overexpression of GhSusA1 increases plant biomass and improves cotton fiber yield and quality. Plant Biotech J 10:301–312
Kim HJ, Murai N, Fang DD, Triplett BA (2011) Functional analysis of Gossypium hirsutum cellulose synthase catalytic subunit 4 promoter in transgenic Arabidopsis and cotton tissues. Plant sci 180(2):323–332
Kim HJ, Triplett BA (2001) Cotton fiber growth in planta and in vitro: models for plant cell elongation and cell wall biogenesis. Plant Physiol 127:1361–1366
Kosambi D (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Kraszewska E (2008) The plant Nudix hydrolase family. Acta Biochim Pol 55(4):663–671
Lande R, Thompson R (1990) Efficiency of marker-assisted selection in the improvement of quantitative traits. Genetics 124(3):743–756
Lee JJ, Woodward AW, Chen ZJ (2007) Gene expression changes and early events in cotton fiber development. Ann Bot (Lond) 100(7):1391–1401
Li FG, Fan GY, Lu CR, Xiao GH, Zou CS, Kohel RJ, Ma ZY, Shang HH, Ma XF, Wu JY, Liang XM, Huang G, Percy RG, Liu K, Yang WH, Chen WB, Du XM, Shi CC, Yuan YL, Ye XL, Liu X, Zhang XY, Liu WQ, Wei HL, Wei SJ, Huang GD, Zhang XL, Zhu SJ, Zhang H, Sun FM, Wang XF, Liang Jie, Wang JH, He Q, Huang LH, Wang J, Cui JJ, Song GL, Wang KB, Xu X, Yu JZ, Zhu YX, Yu SX (2015) Genome sequence of cultivated Upland cotton (Gossypium hirsutumTM-1) provides insights into genome evolution. Nat biotech 33(5):524–530
Li FG, Fan GY, Wang KB, Sun FM, Yuan YL, Song GL, Li Q, Ma ZY, Lu CR, Zou CS, Chen WB, Liang XM, Shang HH, Liu WQ, Shi CC, Xiao GH, Gou CY, Ye WW, Xu X, Zhang XY, Wei HL, Li ZF, Zhang GY, Wang JY, Liu K, Kohel R, Percy RG, Yu JZ, Zhu YX, Wang J, Yu SX (2014) Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet 46:567–572
Li DD, Ruan XM, Zhang J, Wu YJ, Wang XL, Li XB (2013) Cotton plasma membrane intrinsic protein 2 s (PIP2s) selectively interact to regulate their water channel activities and are required for fiber development. New Phytol 199:695–707
Li YL, Sun J, Xia GX (2005) Cloning and characterization of a gene for an LRR receptor-like protein kinase associated with cotton fiber development. Mol Genet Genom 273:217–224.
Li Y, Tu LL, Pettolino FA, Ji SM, Hao J, Yuan DJ, Deng FL, Tan JF, Hu HY, Wang Q, Llewellyn DJ, Zhang XL (2016) GbEXPATR, a species-specific expansin, enhances cotton fiber elongation through cell wall restructuring. Plant biotech J 14(3):951–963.
Liu DX, Zhang J, Liu XY, Wang WW, Liu DJ, Teng ZH, Fang XM, Tan ZY, Tang SY, Yang JH, Zhong JW, Zhang ZS (2016) Fine mapping and RNA-Seq unravels candidate genes for a major QTL controlling multiple fiber quality traits at the T 1 region in upland cotton. BMC genom 17(1):1
Liu BL, Zhu YC, Zhang TZ (2015a) The R3-MYB gene GhCPC negatively regulates cotton fiber elongation. PloS One 10(2): e0116272.
Liu X, Zhao B, Zheng HJ, Hu Y, Lu G, Yang CQ, Chen JD, Chen JJ, Chen DY, Zhang L, Zhou Y, Wang LJ, Guo WZ, Bai YL, Ruan JX, Shangguan XX, Mao YB, Shan CM, Jiang JP, Zhu YQ, Jin L, Kang H, Chen ST, He XL, Wang R, Wang YZ, Chen J, Wang LJ, Yu ST, Wang BY, Wei J, Song SC, Lu XY, Gao ZC, Gu WY, Deng X, Ma D, Wang S, Liang WH, Fang L, Cai CP, Zhu XF, Zhou BL, Chen ZJ, Xu SH, Zhang YG, Wang SY, Zhang TZ, Zhao GP, Chen XY (2015b) Gossypium barbadense genome sequence provides insight into the evolution of extra-long staple fiber and specialized metabolites. Sci Rep 5:1413
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2– ∆∆CT method. Methods 25:402–408
Lv FN, Wang HH, Wang XY, Han LB, Ma YP, Wang S, Feng ZD, Niu XW, Cai CP, Kong ZS, Zhang TZ, Guo WZ (2015) GhCFE1A, a dynamic linker between the ER network and actin cytoskeleton, plays an important role in cotton fiber cell initiation and elongation. J Exp Bot 66(7):1877–1889
Machado A, Wu YR, Yang YM, Llewellyn DJ, Dennis ES (2009) The MYB transcription factor GhMYB25 regulates early fiber and trichome development. Plant J 59:52–62
Monclus R, Leplé JC, Bastien C, Bert PF, Villar M, Marron N, Brignolas F, Jorge V (2012) Integrating genome annotation and QTL position to identify candidate genes for productivity, architecture and water-use efficiency in Populus spp. BMC Plant Biol 12:173
Morris ER, Walker JC (2003) Receptor-like protein kinases: the keys to response. Curr Opin Plant Biol 6(4):339–342
Munis MF, Tu L, Deng F, Tan J, Xu L, Xu S, Long L, Zhang X (2010) A thaumatin-like protein gene involved in cotton fiber secondary cell wall development enhances resistance against Verticillium dahliae and other stresses in transgenic tobacco. Biochem Biophys Res Commu 393:38–44
Niu EL, Cai CP, Zheng YJ, Shang XG, Fang L, Guo WZ (2016) Genome-wide analysis of CrRLK1L gene family in Gossypium and identification of candidate CrRLK1L genes related to fiber development. Mol Genet Genom: 1–18
Ogawa T, Yoshimura K, Miyake H, Ishikawa K, Ito D, Tanabe N, Shigeoka S (2008) Molecular characterization of organelle-type Nudix hydrolases in Arabidopsis. Plant Physiol 148(3):1412–1424
Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin D, Llewellyn D, Showmaker KC, Shu S, Udall J, Yoo MJ, Byers R, Chen W, Doron-Faigenboim A, Duke MV, Gong L, Grimwood J, Grover C, Grupp K, Hu G, Lee TH, Li J, Lin L, Liu T, Marler BS, Page JT, Roberts AW, Romanel E, Sanders WS, Szadkowski E, Tan X, Tang H, Xu C, Wang J, Wang Z, Zhang D, Zhang L, Ashrafi H, Bedon F, Bowers JE, Brubaker CL, Chee PW, Das S, Gingle AR, Haigler CH, Harker D, Hoffmann LV, Hovav R, Jones DC, Lemke C, Mansoor S, ur Rahman M, Rainville LN, Rambani A, Reddy UK, Rong JK, Saranga Y, Scheffler BE, Scheffler JA, Stelly DM, Triplett BA, Van Deynze A, Vaslin MF, Waghmare VN, Walford SA, Wright RJ, Zaki EA, Zhang T, Dennis ES, Mayer KF, Peterson DG, Rokhsar DS, Wang X, Schmutz J (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibers. Nature 492:423–427
Price AH (2006) Believe it or not, QTLs are accurate! Trends Plant Sci 11(5):213–216
Said JI, Song MZ, Wang HT, Lin, ZX, Zhang XL, Fang DD, Zhang JF (2015). A comparative meta-analysis of QTL between intraspecific Gossypium hirsutum and interspecific G. hirsutum × G. barbadense populations. Mol Genet Genom 290(3), 1003–1025
Shan CM, Shangguan XX, Zhao B, Zhang XF, Chao LM, Yang CQ, Wang LJ, Zhu HY, Zeng YD, Guo WZ, Zhou BL, Hu GJ, Guan XY, Chen ZJ, Wendel JF, Zhang TZ, Chen XY (2014) Control of cotton fiber elongation by a homeodomain transcription factor GhHOX3. Nat commu 5:5519
Shang LG, Liang QZ, Wang YM, Wang XC, Wang KB, Abduweli A, Ma LL, Cai SH, Hua JP (2015) Identification of stable QTLs controlling fiber traits properties in multi-environment using recombinant inbred lines in Upland cotton (Gossypium hirsutum L.). Euphytica 205:877–888
Song D, Xi W, Shen J, Bi T, Li L (2011) Characterization of the plasma membrane proteins and receptor-like kinases associated with secondary vascular differentiation in poplar. Plant Mol Biol 76:97–115
Su CF, Wang W, Qiu XM, Yang L, Li S (2013) Fine-mapping a fiber strength QTL QFS-D11-1 on cotton chromosome 21 using introgressed lines. Plant Breed 132:725–730
Sun X, Gong SY, Nie XY, Li Y, Li W, Huang GQ, Li, XB (2015) A R2R3-MYB transcription factor that is specifically expressed in cotton (Gossypium hirsutum) fibers affects secondary cell wall biosynthesis and deposition in transgenic Arabidopsis. Physiol Plant 154:420–432
Sun FD, Zhang JH, Wang SF, Gong WK, Shi YZ, Liu AY, Li JW, Gong JW, Shang HH, Yuan YL (2012) QTL mapping for fiber quality traits across multiple generations and environments in Upland cotton. Mol Breed 30:569–582
Suzuki T, Ishikawa K, Yamashino T, Mizuno T (2002) An Arabidopsis histidine-containing phosphotransfer (HPt) factor implicated in phosphorelay signal transduction: overexpression of AHP2 in plants results in hypersensitiveness to cytokinin. Plant Cell Physiol 43(1):123–129
Suzuki T, Sakurai K, Imamura A, Nakamura A, Ueguchi C, Mizuno T (2000) Compilation and characterization of histidine-containing phosphotransmitters implicated in His-to-Asp phosphorelay in plants: AHP signal transducers of Arabidopsis thaliana. Biosci Biotech Bioch 64(11):2486–2489
Tan ZY, Fang XM, Tang SY, Zhang J, Liu DJ, Teng ZH, Li L, Ni HJ, Zheng FM, Liu DX, Zhang TF, Paterson AH, Zhang ZZ (2015) Genetic map and QTL controlling fiber quality traits in Upland cotton (Gossypium hirsutum L.). Euphytica 203:615–628
Tang SY, Teng ZH, Zhai TF, Fang XM, Liu F, Liu DX, Wang SF, Zhang K, Shao QS, Tan ZY, Paterson AH, Zhang ZZ (2015) Construction of genetic map and QTL analysis of fiber quality traits for Upland cotton (Gossypium hirsutum L.). Euphytica 201:195–213
Tang WX, Tu LL, Yang XY, Tan JF, Deng FL, Hao J, Guo K, Lindsey K, Zhang XL (2014) The calcium sensor GhCaM7 promotes cotton fiber elongation by modulating reactive oxygen species (ROS) production. New Phytol 202:509–520
Van Ooijen JW (2006) JoinMap 4.0: software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen
Van Ooijen J (2009) MapQTL 6, software for the mapping of quantitative trait loci in experimental populations of diploid species. Kyazma BV, Wageningen
Voorrips R (2002) MapChart: software for the graphical presentationof linkage maps and QTLs. J Hered 93:77–78
Walford SA, Wu YR, Llewellyn DJ, Dennis ES (2011) GhMYB25-like: a key factor in early cotton fibre development. Plant J 65:785–797
Walford SA, Wu YR, Llewellyn DJ, Dennis ES (2012) Epidermal cell differentiation in cotton mediated by the homeodomain leucine zipper gene, GhHD-1. Plant J 71:464–478
Wang L, Cook A, Patrick JW, Chen XY, Ruan YL (2014) Silencing the vacuolar invertase gene GhVIN1 blocks cotton fiber initiation from the ovule epidermis, probably by suppressing a cohort of regulatory genes via sugar signaling. Plant J 78:686–696
Wang G, Ellendorff U, Kemp B, Mansfield JW, Forsyth A, Mitchell K, Bastas K, Liu CM, Woods-Tör A, Zipfel C, de Wit PJ, Jones JD, Tör M, Thomma BP (2008) A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol 147(2):503–517
Wang HY, Wang J, Gao P, Jiao GL, Zhao PM, Li Y, Wang GL, Xia GX (2009) Down-regulation ofGhADF1gene expression affects cotton fiber properties. Plant biotech J 7(1):13–23
Wang KB, Wang ZW, Li FG, Ye WW, Wang JY, Song GL, Yue Z, Cong L, Shang HH, Zhu SL, Zou CS, Li Q, Yuan YL, Lu CR, Wei HL, Gou CY, Zheng ZQ, Yin Y, Zhang XY, Liu K, Wang B, Song C, Shi N, Kohel RJ, Percy RG, Yu JZ, Zhu YX, Wang J, Yu SX (2012) The draft genome of a diploid cotton Gossypium raimondii. Nat genet 44(10):1098–1103
Wang FR, Xu ZZ, Sun R, Gong YC, Liu GD, Zhang JX, Wang LM, Zhang CY, Fan SJ, Zhang J (2013a) Genetic dissection of the introgressive genomic components from Gossypium barbadense L. that contribute to improved fiber quality in Gossypium hirsutum L. Mol breed 32(3):547–562
Wang JH, Kucukoglu M, Zhang LB, Chen P, Decker D, Nilsson O, Jones B, Sandberg G, Zheng B (2013b) The Arabidopsis LRR-RLK, PXC1, is a regulator of secondary wall formation correlated with the TDIF-PXY/TDR-WOX4 signaling pathway. BMC Plant Biol 13:94
Xu SL, Rahman A, Baskin TI, Kieber JJ (2008) Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell 20:3065–3079
Yang X, Wang Y, Zhang G, Wang X, Wu L, Ke H, Liu H, Ma Z (2016) Detection and validation of one stable fiber strength QTL on c9 in tetraploid cotton. Mol Genet Genom 291(4):1625–1638
Yang MY, Xu CN, Wang BM, Jia JZ (2001) Effects of plant growth regulators on secondary wall thickening of cotton fibers. Plant Growth Regul 35(3):233–237
Yang ZR, Zhang CJ, Yang XJ, Liu K, Wu ZX, Zhang XY, Zheng Wu, Xun QQ, Liu CL, Lu LL, Yang ZE, Qian YY, Xu ZZ, Li CF, Li J, Li FG (2014) PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytol 203:437–444
Yuan DJ, Tang ZH, Wang MJ, Gao WH, Tu LL, Jin X, Chen LL, He YH, Zhang L, Zhu LF, Li Y, Liang QQ, Lin ZX, Yang XY, Liu N, Jin SX, Lei Y, Ding YH, Li GL, Ruan XA, Ruan YJ, Zhang XL (2015) The genome sequence of SeaIsland cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci Rep 5(6):17662
Zhang TZ, Hu Y, Jiang WK, Fang L, Guan XY, Chen JD, Zhang JB, Saski CA, Scheffler BE, Stelly DM, Hulse-Kemp AM, Wan Q, Liu BL, Liu CX, Wang S, Pan MQ, Wang YK, Wang DW, Ye WX, Chang LJ, Zhang WP, Song QX, Kirkbride RC, Chen XY, Dennis E, Llewellyn DJ, Peterson DG, Thaxton P, Jones DC, Wang Q, Xu XY, Zhang H, Wu HT, Zhou L, Mei GF, Chen SQ, Tian Y, Xiang D, Li XH, Ding J, Zuo QY, Tao LN, Liu YC, Li J, Lin Y, Hui YY, Cao ZS, Cai CP, Zhu XF, Jiang Z, Zhou BL, Guo WZ, Li RQ, Chen ZJ (2015) Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotech 33:531–537
Zhang ZS, Hu MC, Zhang J, Liu DJ, Zheng J, Zhang K, Wang W, Wan Q (2009) Construction of a comprehensive PCR based marker linkage map and QTL mapping for fiber quality traits in Upland cotton (Gossypium hirsutum L.). Mol Breed 24:49–61
Zhang ZS, Xiao YH, Luo M, Li XB, Luo XY, Hou L, Li DM, Pei Y (2005) Construction of a genetic linkage map and QTL analysis of fiber-related traits in Upland cotton (Gossypium hirsutum L.). Euphytica 144:91–99
Zhang K, Zhang J, Ma J, Tang SY, Liu DJ, Teng ZH, Liu DX, Zhang ZZ (2012) Genetic mapping and quantitative trait locus analysis of fiber quality traits using a three-parent composite population in Upland cotton (Gossypium hirsutum L.). Mol breed 29(2):335–348
Zhao J, Gao Y, Zhang Z, Chen TZ, Guo WZ and Zhang TZ. 2013. A receptor-like kinase gene (GbRLK) from Gossypium barbadense enhances salinity and drought-stress tolerance in Arabidopsis. BMC plant Biol 13(1):1
Zhou Y, Zhang ZT, Li M, Wei XZ, Li XJ, Li BY, Li XB (2015) Cotton (Gossypium hirsutum) 14-3-3 proteins participate in regulation of fiber initiation and elongation by modulating brassinosteroid signaling. Plant Biotech J 13(2):269–280
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Nos. 31271770 and 31571720) and National science and technology major projects of China (No. 2016ZX08005003).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The authors declare that this study complies with the current laws of the countries in which the experiments were performed.
Additional information
Communicated by Joshua A Udall.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fang, X., Liu, X., Wang, X. et al. Fine-mapping qFS07.1 controlling fiber strength in upland cotton (Gossypium hirsutum L.). Theor Appl Genet 130, 795–806 (2017). https://doi.org/10.1007/s00122-017-2852-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-2852-1