Abstract
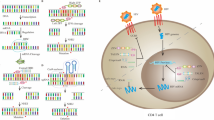
HIV/AIDS remains a major public health issue. In 2014, it was estimated that 36.9 million people are living with HIV worldwide, including 2.6 million children. Since the advent of combination antiretroviral therapy (cART), in the 1990s, treatment has been so successful that in many parts of the world, HIV has become a chronic condition in which progression to AIDS has become increasingly rare. However, while people with HIV can expect to live a normal life span with cART, lifelong medication is required and cardiovascular, renal, liver, and neurologic diseases are still possible, which continues to prompt research for a cure for HIV. Infected reservoir cells, such as CD4+ T cells and myeloid cells, allow persistence of HIV as an integrated DNA provirus and serve as a potential source for the re-emergence of virus. Attempts to eradicate HIV from these cells have focused mainly on the so-called “shock and kill” approach, where cellular reactivation is induced so as to trigger the purging of virus-producing cells by cytolysis or immune attack. This approach has several limitations and its usefulness in clinical applications remains to be assessed. Recent advances in gene-editing technology have allowed the use of this approach for inactivating integrated proviral DNA in the genome of latently infected cells or knocking out HIV receptors. Here, we review this strategy and its potential to eliminate the latent HIV reservoir resulting in a sterile cure of AIDS.


Similar content being viewed by others
References
UNAIDS (2015) http://www.unaids.org/sites/default/files/media_asset/MDG6Report_en.pdf
Lewden C, Chene G, Morlat P et al (2007) HIV-infected adults with a CD4 cell count greater than 500 cells/mm3 on long-term combination antiretroviral therapy reach same mortality rates as the general population. J Acquir Immune Defic Syndr 46:72–77
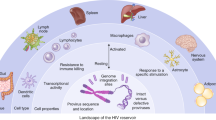
Kulpa DA, Chomont N (2015) HIV persistence in the setting of antiretroviral therapy: when, where and how does HIV hide? J Virus Erad 1:59–66
Saylor D, Dickens AM, Sacktor N et al (2016) HIV-associated neurocognitive disorder—pathogenesis and prospects for treatment. Nat Rev Neurol 12:234–248
Hearps AC, Martin GE, Rajasuriar R, Crowe SM (2014) Inflammatory co-morbidities in HIV + individuals: learning lessons from healthy ageing. Curr HIV/AIDS Rep 11:20–34
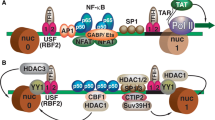
Van Lint C, Bouchat S, Marcello A (2013) HIV-1 transcription and latency: an update. Retrovirology 10:67
Cary DC, Peterlin BM (2016) Targeting the latent reservoir to achieve functional HIV cure. F1000Res. 5
Datta PK, Kaminski R, Hu W et al (2016) HIV-1 latency and eradication: past, present and future. Curr HIV Res 14:431–441
Finzi D, Blankson J, Siliciano JD et al (1999) Latent infection of CD4 + T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 5:512–517
Hütter G, Nowak D, Mossner M et al (2009) Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med 360:692–698
Mummidi S, Ahuja SS, Gonzalez E et al (1998) Genealogy of the CCR5 locus and chemokine system gene variants associated with altered rates of HIV-1 disease progression. Nat Med 4:786–793
de Silva E, Stumpf MP (2004) HIV and the CCR5-Delta32 resistance allele. FEMS Microbiol Lett 241:1–12
Henrich TJ, Hanhauser E, Marty FM et al (2015) Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann Intern Med 161:319–327
Persaud D, Gay H, Ziemniak C et al (2013) Absence of detectable HIV-1 viremia after treatment cessation in an infant. N Engl J Med 369:1828–1835
Hill AL, Rosenbloom DIS, Fud F et al (2014) Predicting the outcomes of treatment to eradicate the latent reservoir for HIV-1. Proc Natl Acad Sci USA 111:13475–13480
Hill AL, Rosenbloom DIS, Siliciano JD et al (2016) Real-time predictions of reservoir size and rebound time during antiretroviral therapy interruption trials for HIV. PLoS Pathog 12:e1005535
Conway JM, Perelson AS (2015) Post-treatment control of HIV infection. Proc Natl Acad Sci USA 112:5467–5472
Conway JM, Perelson AS (2016) Residual viremia in treated HIV + individuals. PLoS Comput Biol 12:e1004677
Shan L, Deng K, Shroff NS et al (2012) Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity 36:491–501
Elliott JH, Wightman F, Solomon A et al (2014) Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog 10:e1004473
Archin NM, Liberty AL, Kashuba AD (2012) Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487:482–486
Archin NM, Bateson R, Tripathy M et al (2014) HIV-1 expression within resting CD4 T-cells following multiple doses of vorinostat. J Infect Dis 210:728–735
Rasmussen TA, Tolstrup M, Brinkmann CR et al (2014) Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV 1:e13–e21
Hamer DH (2004) Can HIV be cured? Mechanisms of HIV persistence and strategies to combat it. Curr HIV Res 2:99–111
Martin AR, Siliciano RF (2016) Progress toward HIV eradication: case reports, current efforts, and the challenges associated with cure. Annu Rev Med 67:215–228
Meinke G, Bohm A, Hauber J, Pisabarro MT, Buchholz F (2016) Cre recombinase and other tyrosine recombinases. Chem Rev (in press)
Gaj T, Guo J, Kato Y, Sirk SJ, Barbas CF (2012) Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat Methods 9:805–807
Kim YG, Li L, Chandrasegaran S (1994) Insertion and deletion mutants of FokI restriction endonuclease. J Biol Chem 269:31978–31982
Wright DA, Li T, Yang B, Spalding MH (2014) TALEN-mediated genome editing: prospects and perspectives. Biochem J 462:15–24
Ousterout DG, Gersbach CA (2016) The development of TALE nucleases for biotechnology. Methods Mol Biol 1338:27–42
Gaj T, Gersbach CA, Barbas CF (2013) ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31:397–405
Sedlak RH, Liang S, Niyonzima N et al (2016) Digital detection of endonuclease mediated gene disruption in the HIV provirus. Sci Rep 6:20064
Mali P, Esvelt KM, Church GM (2013) Cas9 as a versatile tool for engineering biology. Nat Methods 957–963:2013
Doudna JA, Charpentier E (2014) Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096
Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308
Bhaya D, Davison M, Barrangou R (2011) CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet 45:273–997
Buchholz F, Stewart AF (2001) Alteration of Cre recombinase site specificity by substrate-linked protein evolution. Nat Biotechnol 19:1047–1052
Sarkar I, Hauber I, Hauber J, Buchholz F (2007) HIV-1 proviral DNA excision using an evolved recombinase. Science 316:1912–1915
Buchholz F, Hauber J (2011) In vitro evolution and analysis of HIV-1 LTR-specific recombinases. Methods 53:102–109
Buchholz F, Hauber J (2013) Engineered DNA modifying enzymes: components of a future strategy to cure HIV/AIDS. Antiviral Res 97:211–217
Mariyanna L, Priyadarshini P, Hofmann-Sieber H (2012) Excision of HIV-1 proviral DNA by recombinant cell permeable tre-recombinase. PLoS One 7:e31576
Hauber I, Hofmann-Sieber H, Chemnitz J et al (2013) Highly significant antiviral activity of HIV-1 LTR-specific Tre-recombinase in humanized mice. PLoS Pathog 9:e1003587
Karpinski J, Hauber I, Chemnitz J et al (2016) Directed evolution of a recombinase that excises the provirus of most HIV-1 primary isolates with high specificity. Nat Biotechnol 34:401–409
Santiago Y, Chan E, Liu PQ et al (2008) Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci USA 105:5809–5814
Perez EE, Wang J, Miller JC et al (2009) Establishment of HIV-1 resistance in CD4 + T cells by genome editing using zinc-finger nucleases. Nat Biotechnol 26:808–816
Qu X, Wang P, Ding D et al (2013) Zinc-finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells. Nucleic Acids Res 41:7771–7782
Qu X, Wang P, Ding D et al (2014) Zinc finger nuclease: a new approach for excising HIV-1 proviral DNA from infected human T cells. Mol Biol Rep 41:5819–5827
Coakley E, Petropoulos CJ, Whitcomb JM (2005) Assessing chemokine co-receptor usage in HIV. Curr Opin Infect Dis 18:9–15
Berger EA, Doms RW, Fenyö EM et al (1998) A new classification for HIV-1. Nature 391:240
Hardy WD, Gulick RM, Mayer H (2010) Two-year safety and virologic efficacy of maraviroc in treatment-experienced patients with CCR5-tropic HIV-1 infection: 96-week combined analysis of MOTIVATE 1 and 2. J Acquir Immune Defic Syndr 55:558–564
Woollard SM, Kanmogne GD (2015) Maraviroc: a review of its use in HIV infection and beyond. Drug Des Devel Ther 9:5447–5468
Cannon P, June C (2011) Chemokine receptor 5 knockout strategies. Curr Opin HIV AIDS 6:74–79
Holt N, Wang J, Kim K (2010) Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol 28:839–847
Maier DA, Brennan AL, Jiang S (2013) Efficient clinical scale gene modification via zinc finger nuclease-targeted disruption of the HIV co-receptor CCR5. Hum Gene Ther 24:245–258
Li L, Krymskaya L, Wang J (2013) Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Mol Ther 21:1259–1269
Hofer U, Henley JE, Exline CM et al (2013) Pre-clinical modeling of CCR5 knockout in human hematopoietic stem cells by zinc finger nucleases using humanized mice. J Infect Dis 208(Suppl 2):S160–S164
Yi G, Choi JG, Bharaj P (2014) CCR5 gene editing of resting CD4(+) T Cells by transient ZFN expression from HIV envelope pseudotyped nonintegrating lentivirus confers HIV-1 resistance in humanized mice. Mol Ther Nucleic Acids 3:e198
Yao Y, Nashun B, Zhou T et al (2012) Generation of CD34 + cells from CCR5-disrupted human embryonic and induced pluripotent stem cells. Hum Gene Ther 23:238–242
Yuan J, Wang J, Crain K et al (2012) Zinc-finger nuclease editing of human cxcr4 promotes HIV-1 CD4(+) T cell resistance and enrichment. Mol Ther 20:849–859
Didigu CA, Wilen CB, Wang J et al (2014) Simultaneous zinc-finger nuclease editing of the HIV coreceptors CCR5 and CXCR4 protects CD4 + T cells from HIV-1 infection. Blood 123:61–69
Benjamin R, Berges BK, Solis-Leal A et al (2016) TALEN gene editing takes aim on HIV. Hum Genet 135:1059–1070
Strong CL, Guerra HP, Mathew KR et al (2015) Damaging the integrated HIV proviral DNA with TALENs. PLoS One 10:e0125652
Ru R, Yao Y, Yu S et al (2013) Targeted genome engineering in human induced pluripotent stem cells by penetrating TALENs. Cell Regen (Lond) 2:5
Mock U, Riecken K, Berdien B et al (2014) Novel lentiviral vectors with mutated reverse transcriptase for mRNA delivery of TALE nucleases. Sci Rep 4:6409
Mock U, Machowicz R, Hauber I (2015) mRNA transfection of a novel TAL effector nuclease (TALEN) facilitates efficient knockout of HIV co-receptor CCR5. Nucleic Acids Res 43:5560–5571
Fadel HJ, Morrison JH, Saenz DT et al (2014) TALEN knockout of the PSIP1 gene in human cells: analyses of HIV-1 replication and allosteric integrase inhibitor mechanism. J Virol 88:9704–9717
Ebina H, Kanemura Y, Misawa N et al (2015) A high excision potential of TALENs for integrated DNA of HIV-based lentiviral vector. PLoS One 10:e0120047
White MK, Hu W, Khalili K (2015) The CRISPR/Cas9 genome editing methodology as a weapon against human viruses. Discov Med 19:255–262
Khalili K, Kaminski R, Gordon J (2015) Genome editing strategies: potential tools for eradicating HIV-1/AIDS. J Neurovirol 21:310–321
Ebina H, Misawa N, Kanemura Y, Koyanagi Y (2013) Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep 3:2510
Hu W, Kaminski R, Yang F et al (2014) RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc Natl Acad Sci USA 111:11461–11466
Kaminski R, Chen Y, Fischer T et al (2016) Elimination of HIV-1 genomes from human T-lymphoid cells by CRISPR/Cas9 gene editing. Sci Rep 6:22555
Liao HK, Gu Y, Diaz A et al (2015) Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Commun 6:6413
Yin C, Zhang T, Li F et al (2016) Functional screening of guide RNAs targeting the regulatory and structural HIV-1 viral genome for a cure of AIDS. AIDS 30:1163–1174
Zhu W, Lei R, Le Duff Y et al (2015) The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology 12:22
Kaminski R, Bella R, Yin C et al (2016) Excision of HIV-1 DNA by gene editing: a proof-of-concept in vivo study. Gene Ther 23:696
Zhang Y, Yin C, Zhang T et al (2015) CRISPR/gRNA-directed synergistic activation mediator (SAM) induces specific, persistent and robust reactivation of the HIV-1 latent reservoirs. Sci Rep 5:16277
Bialek JK, Dunay GA, Voges M et al (2016) Targeted HIV-1 latency reversal using CRISPR/Cas9-derived transcriptional activator systems. PLoS One 11:e0158294
De Silva Feelixge HS, Stone D et al (2016) Detection of treatment-resistant infectious HIV after genome-directed antiviral endonuclease therapy. Antiviral Res 126:90–98
Wang G, Zhao N, Berkhout B, Das AT (2016) CRISPR-Cas9 can inhibit HIV-1 replication but NHEJ repair facilitates virus escape. Mol Ther 24:522–526
Wang Z, Pan Q, Gendron P et al (2016) CRISPR/Cas9-derived mutations both inhibit HIV-1 replication and accelerate viral escape. Cell Rep 15:481–489
Yoder KE, Bundschuh R (2016) Host double strand break repair generates HIV-1 strains resistant to CRISPR/Cas9. Sci Rep 6:29530
Ueda S, Ebina H, Kanemura Y et al (2016) Anti-HIV-1 potency of the CRISPR/Cas9 system insufficient to fully inhibit viral replication. Microbiol Immunol 60:483–496
Chen X, Gonçalves MA (2016) Engineered viruses as genome editing devices. Mol Ther 24:447–457
Wang D, Mou H, Li S et al (2015) Adenovirus-mediated somatic genome editing of Pten by CRISPR/Cas9 in mouse liver in spite of Cas9-specific immune responses. Hum Gene Ther 26:432–442
Gori JL, Hsu PD, Maeder ML (2015) Delivery and specificity of CRISPR-Cas9 genome editing technologies for human gene therapy. Hum Gene Ther 26:443–451
Choi JG, Dang Y, Abraham S et al (2016) Lentivirus pre-packed with Cas9 protein for safer gene editing. Gene Ther 23:627–633
White MK, Kaminski R, Wollebo H et al (2016) Gene editing for treatment of neurological infections. Neurother 13:547–554
Tebas P, Stein D, Tang WW et al (2014) Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med 370:901–910
Adair JE, Waters T, Haworth KG (2016) Semi-automated closed system manufacturing of lentivirus gene-modified haematopoietic stem cells for gene therapy. Nat Commun 7:13173
Urba WJ, Longo DL (2011) Redirecting T cells. N Engl J Med 365:754–757
Zhen A, Kamata M, Rezek V et al (2015) HIV-specific immunity derived from chimeric antigen receptor-engineered stem cells. Mol Ther 23:1358–1367
Pegu A, Asokan M, Wu L et al (2015) Activation and lysis of human CD4 cells latently infected with HIV-1. Nat Commun 6:8447
Sung JA, Pickeral J, Liu L et al (2015) Dual-Affinity re-targeting proteins direct T cell-mediated cytolysis of latently HIV-infected cells. J Clin Invest 125:4077–4090
Gardner MR, Kattenhorn LM, Kondur HR et al (2015) AAV-expressed eCD4-Ig provides durable protection from multiple SHIV challenges. Nature 519:87–91
Zhen A, Rezek V, Youn C, et al (2016) Stem-cell based engineered immunity against HIV infection in the humanized mouse model. J Vis Exp (in press)
Liu L, Patel B, Ghanem MH et al (2015) Novel CD4-Based Bispecific Chimeric Antigen Receptor Designed for Enhanced Anti-HIV Potency and Absence of HIV Entry Receptor Activity. J Virol 89:6685–6694
Sahu GK, Sango K, Selliah N, et al (2013) Anti-HIV designer T cells progressively eradicate a latently infected cell line by sequentially inducing HIV reactivation then killing the newly gp120-positive cells. Virology 446:268–275
Wang CX, Cannon PM (2016) The clinical applications of genome editing in HIV. Blood 127:2546–2552
Kishida T, Ejima A, Mazda O (2016) Specific destruction of HIV proviral p17 gene in T lymphoid cells achieved by the genome editing technology. Front Microbiol 7:1001
Kang H, Minder P, Park MA et al (2015) CCR5 disruption in induced pluripotent stem cells using CRISPR/Cas9 provides selective resistance of immune cells to CCR5-tropic HIV-1 virus. Mol Ther Nucleic Acids 4:e268
Randhawa S, Cho BS, Ghosh D et al (2016) Effects of pharmacological and genetic disruption of CXCR4 chemokine receptor function in B-cell acute lymphoblastic leukaemia. Br J Haematol 174:425–436
Hou P, Chen S, Wang S, et al (2015) Genome editing of CXCR4 by CRISPR/cas9 confers cells resistant to HIV-1 infection. Sci Rep 5:15577
Schumann K, Lin S, Boyer E et al (2015) Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci USA 112:10437–10442
Holt N, Wang J, Kim K eel al (2010) Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol 28:839–847
Wang W, Ye C, Liu J et al (2014) CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS One 9:e115987
Acknowledgements
We thank the past and present members of the Department of Neuroscience for their continued support and insightful discussions. We also acknowledge the intellectual contributions of the Katz School of Medicine at Temple University Comprehensive NeuroAIDS Center (Basic Science Cores I and II) (NIH P30MH092177). We are grateful to Cynthia Papaleo for editorial assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Khalili, K., White, M.K. & Jacobson, J.M. Novel AIDS therapies based on gene editing. Cell. Mol. Life Sci. 74, 2439–2450 (2017). https://doi.org/10.1007/s00018-017-2479-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-017-2479-z