Abstract
Despite the growing understanding of the mechanisms of carcinogenesis, cancers of the central nervous system are usually associated with unfavorable prognosis. The use of an appropriate molecular marker may improve the treatment outcome by allowing early diagnosis and treatment susceptibility monitoring. Since methylation of tumor-derived DNA can be detected in the serum of cancer patients, this makes DNA methylation-based biomarkers one of the most promising diagnostic strategies. In this study, the methylation profiles of MGMT, RASSF1A, p15INK4B, and p14ARF genes were evaluated in serum free-circulating DNA and the corresponding tumor tissue in a group of 33 primary or metastatic central nervous system cancer patients. Gene promoter methylation was assessed using methylation-specific polymerase chain reaction (PCR). All the tested genes were found to be methylated to a different extent in both serum and tumor samples. In comparison to metastatic brain tumor patients, the patients with glial tumors were characterized by a higher frequency of gene hypermethylation. The hypermethylation of RASSF1A differentiated primary from metastatic brain cancers. Moreover, the gene methylation profiles observed in serum, in most cases, matched the methylation profiles detected in paired tumor samples.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
It is widely accepted that the deregulation of cellular homeostasis observed in cancer cells may be a consequence of the aberrations in the epigenetic information besides genetic mutations (Kulis and Esteller 2010). Epigenetic mechanisms are responsible for the regulation of gene expression and their frequent changes in carcinogenesis are especially attractive for investigation because of their potential applicability in both tumor diagnostics and therapy. The hypermethylation of CpG islands in the promoter regions of tumor suppressor genes, which leads to their silencing, can be found at all stages of carcinogenesis. The profile of DNA methylation changes is claimed to be relatively specific for tumor (sub)type, which makes it a promising biomarker for disease detection and stratification. Evidence shows that it may be also used for both prognostic and predictive purposes (Paluszczak and Baer-Dubowska 2006).
Central nervous system (CNS) cancers are usually highly malignant, chemo- and radio-resistant, and difficult to cure by surgical resection. The most common brain cancers of adults are gliomas, with glioblastoma (GBM) being the most common and most aggressive. Despite advances in neuro-imaging, neurosurgery, and chemotherapy, the survival time of GBM patients very rarely exceeds 2 years, and the median survival time is only about 12 months (Johnson et al. 2012). The observation that the hypermethylation of MGMT may be a good predictive factor of the response of CNS cancer patients to alkylating anti-cancer drugs, especially temozolomide, led to attempts of clinical application of epigenetic diagnostics (Esteller et al. 2000; Hegi et al. 2004, 2005). The specificity of the predictive testing of MGMT methylation is, however, sometimes questioned, since this drug resistance phenotype may also depend on other molecular changes (McEllin et al. 2010; Zhang et al. 2010). The elaboration of a diagnostic gene panel including MGMT could potentially improve clinical validity. Apart from MGMT, DNA methylation changes in CNS tumors also affect other genes, especially tumor suppressors, such as RASSF1A, p15INK4B, or p14ARF (Lorente et al. 2009; Mulholland et al. 2012; Riemenschneider et al. 2010; Wolter et al. 2001). Their silencing is one of the mechanisms which finally lead to uncontrolled cell proliferation and evading apoptosis and, subsequently, to the acquisition of an aggressive phenotype. Hypermethylation of those genes was found to be involved not only in the carcinogenesis of the CNS tissues, but also breast (Buyru et al. 2009; Fiegl et al. 2005; Sharma et al. 2007) or lung (Furonaka et al. 2004; Ramirez et al. 2003) and in malignant melanoma (Freedberg et al. 2008; Tellez et al. 2009). Such solid tumors often metastasize to the brain and it is thought that DNA methylation changes may be responsible for the acquisition of cerebral metastatic potential by those cells. Metastases are the most common tumors of the CNS, and lung carcinoma, melanoma, and breast carcinoma are the primary tumors most frequently involved in brain invasion (Gavrilovic and Posner 2005; Gonzalez-Gomez et al. 2004).
Because of the limitations in the accessibility of CNS tumor tissue for diagnostic purposes, other sources of tumor-derived DNA are in high demand. There is evidence that significantly higher levels of free-circulating DNA are present in the serum of solid tumor patients and it is believed that most of this DNA is derived from tumor cells (Fiegl et al. 2005; Fleischhacker and Schmidt 2007). Although CNS tumors may shed free DNA into the extracellular space at the same rate as systematic tumors, several anatomic and physiologic differences make it uncertain as to how much of this DNA may reach systematic circulation. Primary CNS tumors are confined to the cranial vault, where their extracellular space drains largely into the cerebrospinal fluid (CSF), which, following circulation, will eventually clear into the bloodstream. This sink effect of the CSF may substantially dilute the amount of detectable circulating nucleic acids in the blood samples of patients with CNS tumors (Lavon et al. 2010).
So far, only a few studies have evaluated the levels of free-circulating DNA in CNS neoplasms in the context of the detection of gene promoter methylation (Lavon et al. 2010; Wakabayashi et al. 2009; Weaver et al. 2006). Moreover, different protocols for sample collection and circulating DNA assessment were used in these studies, and a limited number of gene promoters was evaluated. Since the quantity of free-circulating DNA found in serum is low, the use of an appropriate method of DNA extraction is of high importance. As a result of the comparison of different methods of isolation of DNA from the serum of colorectal cancer patients, Fong et al. (2009) proposed that the sodium iodide protocol is the best option.
In this study, we compared the profile of aberrant methylation of MGMT, RASSF1A, p15INK4B, and p14ARF genes in serum free-circulating DNA and corresponding tumor tissue in a group of CNS cancer patients. MGMT, the O6-methylguanine-DNA methyltransferase gene, is located at chromosome 10q26 and encodes a DNA repair enzyme that can abrogate the effects of alkylating chemotherapy, such as temozolomide. RASSF1A encodes a Ras association domain family member 1 protein, which interacts with DNA repair protein XPA and is frequently inactivated by hypermethylation of its promoter region. p15INK4B, cyclin-dependent kinase inhibitor 2B gene, and p14ARF (alternative transcript of the INK4b-ARF-INK4a locus located on human chromosome 9p21) are tumor suppressors inactivated in a variety of cancers. p15INK4B encodes a cyclin-dependent kinase inhibitor, whereas p14ARF functions as a stabilizer of p53. Those genes were selected based on available information concerning the role of their methylation in brain carcinogenesis. In the majority of cases, gene methylation profiles detected in free-circulating DNA reflected methylation profiles observed in tumor tissue samples.
Patients and methods
Patient demographics
Thirty-three newly diagnosed, previously untreated patients under the care of the Department of Neurosurgery and Neurotraumatology at the Poznań University of Medical Sciences were included in the study (18 men and 15 women). Seventeen patients were diagnosed with brain tumor of glial origin: primary or recurrent glioblastoma (7 patients), astrocytoma (8), or gliosarcoma (2). The remaining 16 patients suffered from either meningiomas (6 patients) or metastatic CNS cancer (10). The median age was 57.9 years (ranging from 35 to 75 years). Serum samples were obtained from all patients, while paired tumor tissue specimens were available for 16 patients. The clinical characteristics of the study group, including sex, age, and clinical diagnoses, are presented in Tables 1 and 2. The research was approved by the local Ethics Committee at the Poznań University of Medical Sciences.
DNA isolation
DNA from the tumor tissue was isolated using the GeneMATRIX Tissue DNA Purification Kit (EURx, Gdańsk, Poland), according to the manufacturer’s protocol. Circulating tumor-derived DNA was isolated from serum using the sodium iodide/glycogen method. Briefly, enzyme reaction solution, containing 1.43 % SDS, 7.15 mM EDTA, 14.3 mM Tris, (pH 8.0), and Proteinase K, was added to serum and the mixture was incubated at 56 °C for 2 h. Subsequently, glycogen solution (Fermentas, Burlington, Canada) was added, followed by the addition of 9M sodium iodide and an equal volume of isopropanol. After a 15-min incubation, the sample was centrifuged (10,000 × g for 20 min) and the pellet was washed with 45 % isopropanol and centrifuged. Washing was repeated with 75 % ethanol. After the final centrifugation, the pellet was dried and dissolved in Tris-EDTA buffer. The concentration and quality of DNA samples was assessed using a NanoDrop spectrophotometer. The mean recovery of total DNA from 3 ml of serum was 8.62 μg (mean concentration of 43.1 ng/μl, with an elution volume of 200 μl).
DNA methylation analysis
The assessment of the methylation status of MGMT, RASSF1A, p15INK4B, and p14ARF was performed using methylation-specific polymerase chain reaction (PCR) (Herman et al. 1996). Completely methylated control DNA (CpG Methylated HeLa Genomic DNA, New England Biolabs, UK) and completely unmethylated control DNA (EpiTect Control DNA, Qiagen, Germany) served as positive and negative controls, respectively. Additionally, leukocytes from healthy volunteers as well as a DNA sample extracted from a brain fragment of a patient with brain hematoma served as normal control samples for DNA methylation analysis. They showed no detectable hypermethylation of the genes analyzed.
Bisulfite modification of 500 ng DNA was performed using the EZ DNA Methylation Kit (Zymo Research, Orange, CA, USA), following manufacturer’s instructions. Methylation-specific PCR assay was carried out in a MyCycler Thermal Cycler (Bio-Rad). Primer oligonucleotides were obtained from Oligo.pl (Warsaw, Poland). Primer sequences for all the analyzed genes and reaction conditions were chosen based on previously published protocols (Dong et al. 2005; Herman et al. 1996; Pizzi et al. 2005; Table 3). All primer pairs were targeted towards gene promoter-associated CpG islands. PCR reactions were carried out in a volume of 18 μl using FastStart Taq DNA Polymerase (Roche, Germany) as follows: after an initial step of heat denaturation at 94 °C for 4 min, 38 cycles of 94 °C for 30 s, appropriate annealing temperature for 30 s, and 72 °C for 45 s were carried out. Final elongation was carried out at 72 °C for 7 min. PCR products were separated by 2 % agarose gel electrophoresis in the presence of ethidium bromide and visualized under UV illumination.
Statistical analysis
Data were analyzed using Fisher’s exact test, with p ≤ 0.05 being considered as statistically significant.
Results
The applied protocol of DNA isolation from serum yielded an amount sufficient for the assessment of gene hypermethylation in CNS cancer patients. Methylation of at least one promoter was demonstrated in 12 out of 17 (70.58 %) serum samples from the glial tumor group, where 5 patients had one, 6 patients had two, and 1 patient had three promoters methylated, respectively (Table 1). In this group, MGMT, RASSF1A, p15INK4B, and p14ARF were methylated in 3/17 (17.65 %), 8/17 (47.06 %), 2/17 (11.76 %), and 7/17 (41.18 %) cases, respectively. At least one promoter was methylated in the serum of 7 out of 10 metastatic CNS tumor patients and 3 out of 6 meningothelial tumor patients (Table 2). MGMT was methylated in the circulating tumor-derived DNA from only one meningioma patient, RASSF1A methylation was found in two meningiomas and three metastatic tumors, and the methylation of both p15INK4B and p14ARF was detected in one meningioma and three metastatic tumors, respectively.
Apart from serum, samples of the corresponding tumor tissue were available for 9 patients with glial and 7 patients with non-glial tumors. The comparative analysis of DNA methylation between these two sample sources is presented in Table 4. The concordance of gene hypermethylation results was relatively high for all the analyzed genes. Among all the patients, 13 discordant results (20.3 %) were observed. These were mostly false-negative cases (in respect to serum analysis). Interestingly, false-positive results were observed mostly (75 %) in the group of metastatic cancers.
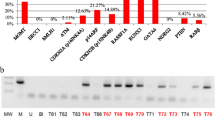
No statistical differences in the frequency of gene hypermethylation between glial and non-glial cancer patients were found when it comes to neither serum nor tumor tissue, except for RASSF1A. In this case, gene promoter methylation was significantly more frequent in the samples of glial tumors in comparison to metastatic CNS cancers (p = 0.0406) (Fig. 1).
Discussion
In the past few years, much data have been gathered showing that cancers of different origins release DNA into the bloodstream (Fiegl et al. 2005; Fleischhacker and Schmidt 2007). Even though the data concerning solid tumors, such as lung, colon, or breast, showed the presence of tumor-derived DNA in the blood, a limited number referred to CNS cancers. The classical view of the brain as an isolated environment, with the blood–brain barrier being impenetrable to large molecules such as DNA, suggested possible obstacles in finding methylation biomarkers in the blood of those patients. However, a recent verification of this hypothesis showed that tumor-derived DNA is present in the serum of glioma patients and that its analysis is eligible for biomarker purposes (Lavon et al. 2010; Wakabayashi et al. 2009; Weaver et al. 2006). Our observations confirm the findings that circulating tumor-derived DNA is present in the serum of CNS cancer patients. Additionally, we verified the utility of the sodium iodide method in yielding DNA of appropriate quantity and quality for further methylation detection, as suggested previously in the case of colorectal tumors (Fong et al. 2009). Because of poor treatment outcomes in the group of brain cancer patients, laboratory tests based on methylation analysis of tumor-derived free-circulating DNA may be potentially useful in patient prognosis and the choice of the best suited therapy regimen (Weaver et al. 2006).
In the present study, we analyzed the promoter methylation of MGMT, RASSF1A, p15INK4B, and p14ARF. It has been proven that these genes play a role in the pathogenesis of CNS cancers, where promoter hypermethylation is one of the mechanisms of their silencing. For example, it was suggested that p14ARF hypermethylation and MGMT hypermethylation constitute distinct molecular pathways of astrocytoma progression, and it is hypothesized that these pathways could differ in biological behavior and clinical outcome (Watanabe et al. 2007). From the diagnostic point of view, MGMT hypermethylation is an important predictive marker of good response to alkylating chemotherapy in GBM. Interestingly, some reports support the thesis that serum-based MGMT methylation analysis offers a promising alternative to a tumor-based approach (Balańa et al. 2011). Although the assessment of MGMT hypermethylation is a valuable marker in the prediction of the response of GBM patients to temozolomide treatment (Silber et al. 2012), some studies indicate that its diagnostic performance is not satisfactory and other methylation-based markers are proposed, e.g., PTEN or hMLH1 (McEllin et al. 2010; Plumb et al. 2000). Therefore, further studies aiming at finding new relevant markers are necessary in order to improve the effectiveness of therapy of CNS cancer patients.
Several studies showed that serum might be a source for circulating DNA. In contrast to plasma, serum contains significantly higher amounts of DNA, with a low level of contaminating extraneous DNA released from leukocytes (Umetani et al. 2006). Thus, we first analyzed the methylation of the selected panel of genes in serum samples obtained from patients with CNS cancers of glial or non-glial (mostly metastatic) origin. Overall, the rate of hypermethylation of the tested genes, besides p15INK4B, was higher in the group of patients with tumors of glial origin. We report frequent methylation of RASSF1A in circulating tumor-derived DNA found in the serum of patients with both primary as well as metastatic CNS tumors. This finding is in agreement with published data, which show the involvement of RASSF1A epigenetic silencing in CNS carcinogenesis (Lorente et al. 2009). When it comes to serum RASSF1A methylation-positive cases of brain tumor patients, similar results were obtained in the study of Dammann et al. (2005). The second gene which also showed frequent hypermethylation in the studied patient group was p14ARF, and recent data suggest that methylation in the promoter region of p14ARF may be used as a biomarker for the diagnosis of gliomas (He et al. 2011). We observed an intermediate frequency of MGMT methylation in the serum of patients with glial tumors (17.65 %), while it was low (6.25 %) in the serum of non-glial tumor patients. The percentage of CNS cancer cases where methylated MGMT was detected in the serum ranged from 20 to 50 % in other reports (Lavon et al. 2010; Weaver et al. 2006). Finally, p15INK4B was observed to be methylated with moderate frequency in the serum of cancers of both glial origin (11.76 %) and in the group consisting of non-glial tumor patients (25 %).
Of 33 serum samples under analysis, 16 had paired frozen tissues available for assessment. The frequencies of gene hypermethylation in tumor samples were similar to those detected in serum with some exceptions, resulting probably from the different number of patients in the groups. The corresponding values were in agreement with previously published data concerning the methylation of MGMT (Cecener et al. 2012; Mellai et al. 2012), RASSF1A (Dammann et al. 2005; Gao et al. 2004; Horiguchi et al. 2003), p14ARF (He et al. 2011; Nakamura et al. 2011), or p15INK4B (Watanabe et al. 2007; Yin et al. 2002) in CNS tumor cells, especially when taking into account the glial tumor group. Moreover, for the first time, we propose that RASSF1A hypermethylation may differentiate primary from metastatic CNS cancers.
The comparison of the results obtained for paired serum and tumor samples allowed the conclusion that the global concordance of results between these two sample sources is fairly high (Table 4). It is generally believed that serum-based marker analysis shows higher specificity (which is reflected by the rate of false-positive results) than sensitivity (reflected by the rate of false-negative results) of detection (Laird 2003). This assumption is confirmed also in our studies. Thus, in general, biomarker methylation was more frequently found in DNA from tumor than from serum, similar to that reported by others (Board et al. 2008). Also, in the studies of Weaver et al. (2006) and Lavon et al. (2010), 40 to 50 % of patients were characterized by the hypermethylation of MGMT in tumor samples, while only half of them showed paired MGMT methylation in free-circulating serum DNA. This can possibly be explained by the fragmentation of circulating tumor-derived DNA. Interestingly, most cases of false-positive gene methylation results were detected in the group of metastatic CNS cancer patients, while false-negative results were more frequently found in glial tumor patients. The possible explanation of this phenomenon is that the methylation markers detected in the serum of metastatic tumor patients can come from DNA originating in the primary tumor site and the metastatic cells creating the CNS tumor may not fully reflect the DNA methylation profile characterizing cells in the primary tumor tissue.
Conclusions
In conclusion, our study seems to confirm the previous findings (Lavon et al. 2010; Wakabayashi et al. 2009; Weaver et al. 2006) that the blood–brain barrier does not interfere significantly with the leakage of tumor-specific DNA into the global circulation, which makes the detection of methylation biomarkers in the serum of CNS cancer patients possible. Gene methylation in the tumor was accompanied by its methylation in paired serum samples in only half of positive cases. The sensitivity of the assay is, therefore, not satisfactory, and further research is necessary in order to establish a marker panel with optimal clinical performance. The results, however, illustrate that the circulating tumor-derived DNA harbors the methylation markers found in the originating tumor tissue and is eligible for clinical biomarker assessment. Additionally, an important novel finding is that metastatic CNS cancers are characterized by a different spectrum of epigenetic changes than primary glial tumors, with RASSF1A hypermethylation differentiating between these two groups. Our work further supports the search for epigenetic CNS tumor markers.
References
Balańa C, Carrato C, Ramírez JL, Cardona AF, Berdiel M, Sánchez JJ, Tarón M, Hostalot C, Musulen E, Ariza A, Rosell R (2011) Tumour and serum MGMT promoter methylation and protein expression in glioblastoma patients. Clin Transl Oncol 13:677–685
Board RE, Knight L, Greystoke A, Blackhall FH, Hughes A, Dive C, Ranson M (2008) DNA methylation in circulating tumour DNA as a biomarker for cancer. Biomark Insights 2:307–319
Buyru N, Altinisik J, Ozdemir F, Demokan S, Dalay N (2009) Methylation profiles in breast cancer. Cancer Invest 27:307–312
Cecener G, Tunca B, Egeli U, Bekar A, Tezcan G, Erturk E, Bayram N, Tolunay S (2012) The promoter hypermethylation status of GATA6, MGMT, and FHIT in glioblastoma. Cell Mol Neurobiol 32:237–244
Dammann R, Schagdarsurengin U, Seidel C, Strunnikova M, Rastetter M, Baier K, Pfeifer GP (2005) The tumor suppressor RASSF1A in human carcinogenesis: an update. Histol Histopathol 20:645–663
Dong SM, Lee EJ, Jeon ES, Park CK, Kim KM (2005) Progressive methylation during the serrated neoplasia pathway of the colorectum. Mod Pathol 18:170–178
Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG (2000) Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 343:1350–1354
Fiegl H, Millinger S, Mueller-Holzner E, Marth C, Ensinger C, Berger A, Klocker H, Goebel G, Widschwendter M (2005) Circulating tumor-specific DNA: a marker for monitoring efficacy of adjuvant therapy in cancer patients. Cancer Res 65:1141–1145
Fleischhacker M, Schmidt B (2007) Circulating nucleic acids (CNAs) and cancer—a survey. Biochim Biophys Acta 1775:181–232
Fong SL, Zhang JT, Lim CK, Eu KW, Liu Y (2009) Comparison of 7 methods for extracting cell-free DNA from serum samples of colorectal cancer patients. Clin Chem 55:587–589
Freedberg DE, Rigas SH, Russak J, Gai W, Kaplow M, Osman I, Turner F, Randerson-Moor JA, Houghton A, Busam K, Timothy Bishop D, Bastian BC, Newton-Bishop JA, Polsky D (2008) Frequent p16-independent inactivation of p14ARF in human melanoma. J Natl Cancer Inst 100:784–795
Furonaka O, Takeshima Y, Awaya H, Ishida H, Kohno N, Inai K (2004) Aberrant methylation of p14(ARF), p15(INK4b) and p16(INK4a) genes and location of the primary site in pulmonary squamous cell carcinoma. Pathol Int 54:549–555
Gao Y, Guan M, Su B, Liu W, Xu M, Lu Y (2004) Hypermethylation of the RASSF1A gene in gliomas. Clin Chim Acta 349:173–179
Gavrilovic IT, Posner JB (2005) Brain metastases: epidemiology and pathophysiology. J Neurooncol 75:5–14
Gonzalez-Gomez P, Bello MJ, Alonso ME, Amiñoso C, Lopez-Marin I, De Campos JM, Isla A, Gutierrez M, Rey JA (2004) Promoter methylation status of multiple genes in brain metastases of solid tumors. Int J Mol Med 13:93–98
He J, Qiao JB, Zhu H (2011) p14ARF promoter region methylation as a marker for gliomas diagnosis. Med Oncol 28:1218–1224
Hegi ME, Diserens AC, Godard S, Dietrich PY, Regli L, Ostermann S, Otten P, Van Melle G, de Tribolet N, Stupp R (2004) Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res 10:1871–1874
Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R (2005) MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352:997–1003
Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB (1996) Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 93:9821–9826
Horiguchi K, Tomizawa Y, Tosaka M, Ishiuchi S, Kurihara H, Mori M, Saito N (2003) Epigenetic inactivation of RASSF1A candidate tumor suppressor gene at 3p21.3 in brain tumors. Oncogene 22:7862–7865
Johnson DR, Ma DJ, Buckner JC, Hammack JE (2012) Conditional probability of long-term survival in glioblastoma: a population-based analysis. Cancer 118:5608–5613. doi:10.1002/cncr.27590
Kulis M, Esteller M (2010) DNA methylation and cancer. Adv Genet 70:27–56
Laird PW (2003) The power and the promise of DNA methylation markers. Nat Rev Cancer 3:253–266
Lavon I, Refael M, Zelikovitch B, Shalom E, Siegal T (2010) Serum DNA can define tumor-specific genetic and epigenetic markers in gliomas of various grades. Neuro Oncol 12:173–180
Lorente A, Mueller W, Urdangarín E, Lázcoz P, Lass U, von Deimling A, Castresana JS (2009) RASSF1A, BLU, NORE1A, PTEN and MGMT expression and promoter methylation in gliomas and glioma cell lines and evidence of deregulated expression of de novo DNMTs. Brain Pathol 19:279–292
McEllin B, Camacho CV, Mukherjee B, Hahm B, Tomimatsu N, Bachoo RM, Burma S (2010) PTEN loss compromises homologous recombination repair in astrocytes: implications for glioblastoma therapy with temozolomide or poly(ADP-ribose) polymerase inhibitors. Cancer Res 70:5457–5464
Mellai M, Monzeglio O, Piazzi A, Caldera V, Annovazzi L, Cassoni P, Valente G, Cordera S, Mocellini C, Schiffer D (2012) MGMT promoter hypermethylation and its associations with genetic alterations in a series of 350 brain tumors. J Neurooncol 107:617–631
Mulholland S, Pearson DM, Hamoudi RA, Malley DS, Smith CM, Weaver JM, Jones DT, Kocialkowski S, Bäcklund LM, Collins VP, Ichimura K (2012) MGMT CpG island is invariably methylated in adult astrocytic and oligodendroglial tumors with IDH1 or IDH2 mutations. Int J Cancer 131:1104–1113
Nakamura M, Watanabe T, Klangby U, Asker C, Wiman K, Yonekawa Y, Kleihues P, Ohgaki H (2011) p14ARF deletion and methylation in genetic pathways to glioblastomas. Brain Pathol 11:159–168
Paluszczak J, Baer-Dubowska W (2006) Epigenetic diagnostics of cancer—the application of DNA methylation markers. J Appl Genet 47:365–375
Pizzi S, Azzoni C, Bottarelli L, Campanini N, D’Adda T, Pasquali C, Rossi G, Rindi G, Bordi C (2005) RASSF1A promoter methylation and 3p21.3 loss of heterozygosity are features of foregut, but not midgut and hindgut, malignant endocrine tumours. J Pathol 206:409–416
Plumb JA, Strathdee G, Sludden J, Kaye SB, Brown R (2000) Reversal of drug resistance in human tumor xenografts by 2′-deoxy-5-azacytidine-induced demethylation of the hMLH1 gene promoter. Cancer Res 60:6039–6044
Ramirez JL, Sarries C, de Castro PL, Roig B, Queralt C, Escuin D, de Aguirre I, Sanchez JM, Manzano JL, Margelí M, Sanchez JJ, Astudillo J, Taron M, Rosell R (2003) Methylation patterns and K-ras mutations in tumor and paired serum of resected non-small-cell lung cancer patients. Cancer Lett 193:207–216
Riemenschneider MJ, Hegi ME, Reifenberger G (2010) MGMT promoter methylation in malignant gliomas. Target Oncol 5:161–165
Sharma G, Mirza S, Prasad CP, Srivastava A, Gupta SD, Ralhan R (2007) Promoter hypermethylation of p16INK4A, p14ARF, CyclinD2 and Slit2 in serum and tumor DNA from breast cancer patients. Life Sci 80:1873–1881
Silber JR, Bobola MS, Blank A, Chamberlain MC (2012) O(6)-methylguanine-DNA methyltransferase in glioma therapy: promise and problems. Biochim Biophys Acta 1826:71–82
Tellez CS, Shen L, Estécio MR, Jelinek J, Gershenwald JE, Issa JP (2009) CpG island methylation profiling in human melanoma cell lines. Melanoma Res 19:146–155
Umetani N, Hiramatsu S, Hoon DS (2006) Higher amount of free circulating DNA in serum than in plasma is not mainly caused by contaminated extraneous DNA during separation. Ann N Y Acad Sci 1075:299–307
Wakabayashi T, Natsume A, Hatano H, Fujii M, Shimato S, Ito M, Ohno M, Ito S, Ogura M, Yoshida J (2009) p16 promoter methylation in the serum as a basis for the molecular diagnosis of gliomas. Neurosurgery 64:455–461
Watanabe T, Katayama Y, Yoshino A, Yachi K, Ohta T, Ogino A, Komine C, Fukushima T (2007) Aberrant hypermethylation of p14ARF and O6-methylguanine-DNA methyltransferase genes in astrocytoma progression. Brain Pathol 17:5–10
Weaver KD, Grossman SA, Herman JG (2006) Methylated tumor-specific DNA as a plasma biomarker in patients with glioma. Cancer Invest 24:35–40
Wolter M, Reifenberger J, Blaschke B, Ichimura K, Schmidt EE, Collins VP, Reifenberger G (2001) Oligodendroglial tumors frequently demonstrate hypermethylation of the CDKN2A (MTS1, p16INK4a), p14ARF, and CDKN2B (MTS2, p15INK4b) tumor suppressor genes. J Neuropathol Exp Neurol 60:1170–1180
Yin D, Xie D, Hofmann WK, Miller CW, Black KL, Koeffler HP (2002) Methylation, expression, and mutation analysis of the cell cycle control genes in human brain tumors. Oncogene 21:8372–8378
Zhang J, Stevens MF, Laughton CA, Madhusudan S, Bradshaw TD (2010) Acquired resistance to temozolomide in glioma cell lines: molecular mechanisms and potential translational applications. Oncology 78:103–114
Acknowledgments
This work was supported by the Polish National Science Centre grant no. NN405 683240.
Conflict of interest
The authors declare no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Majchrzak-Celińska, A., Paluszczak, J., Kleszcz, R. et al. Detection of MGMT, RASSF1A, p15INK4B, and p14ARF promoter methylation in circulating tumor-derived DNA of central nervous system cancer patients. J Appl Genetics 54, 335–344 (2013). https://doi.org/10.1007/s13353-013-0149-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-013-0149-x