Abstract
Purpose
Although meningiomas are common central nervous system tumors, the biomarkers allowing early diagnosis and progression are still needed. The aim of this study was to evaluate the methylation status of 12 cancer-related genes, namely ERCC1, hMLH1, ATM, CDKN2B (p15INK4B), p14ARF, CDKN2A (p16INK4A), RASSF1A, RUNX3, GATA6, NDRG2, PTEN, and RARβ, in 44 meningioma samples of WHO grade I and II.
Methods
All genes were analyzed using methylation-specific polymerase chain reaction, while pyrosequencing (PSQ) was used to study NDRG2 promoter methylation.
Results
The most frequently methylated genes in both types of meningiomas were p14ARF, RASSF1A, and p15INK4B. RUNX3, GATA6, and p16INK4A were methylated to a lesser extent, whereas ATM and RARβ were found to be methylated in a marginal number of patients. The ERCC1, hMLH1, NDRG2, and PTEN genes were unmethylated in all cases. Although tumors of the same grade according to WHO criteria had different genes methylated, the number of methylated genes for each individual patient was low. RUNX3 methylation significantly correlated with meningioma WHO grade, therefore, can be considered as a potential indicator of tumor aggressiveness. The sequence of NDRG2 chosen for PSQ analysis was found methylated in the majority of meningiomas; however, the methylation level was only slightly elevated as compared to non-cancerous brain.
Conclusions
Overall, the results of this study confirm that DNA methylation plays an important role in the pathogenesis of meningiomas. Further investigations, particularly concerning RUNX3 methylation, are necessary in order to assess the clinical usefulness of the methylation analysis of the studied genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The most common tumor site in the central nervous system is the meninges (35 %) and meningiomas, with the incidence rate of 7.22 per 100,000 persons, are the most common primary brain tumors (Dolecek et al. 2012). Incidence rate for tumors of the meninges is 2.2 times greater in females as compared to males, and the peak incidence occurs in the sixth and seventh decade of life (Dolecek et al. 2012; Lamszus 2004). They are classified according to the World Health Organization (WHO) grading system into grade I, II, or III tumors, and around 80 % of meningiomas are grade I tumors (benign), 10–15 % are grade II (atypical), and 2–5 % are grade III (anaplastic/malignant) (Cahill and Claus 2011). Grade I tumors are mostly curable by surgical resection and have good prognosis; however, some populations of meningiomas with benign histological profiles show malignant behavior. The reason for this inconsistency is yet to be elucidated, and the search for novel diagnostic, prognostic, and predictive biomarkers which could help in meningioma patients stratification and treatment is therefore needed. In addition to alterations in the DNA sequence, epigenetic modifications are closely linked to cancer initiation and progression. Aberrant DNA methylation is one of the major types of epigenetic modifications in cancer (Paluszczak and Baer-Dubowska 2006). Changes in DNA methylation appearing during early stages of malignancy are potentially reversible and in some cases correlate well with tumor characteristics. For instance, TIMP3 inactivation as a result of gene promoter methylation appears to be involved in meningioma progression as it is associated with more aggressive, high-grade meningioma phenotype (Barski et al. 2010). Recently, three genome-wide methylation analyses of benign, atypical, and malignant meningiomas were performed (Kishida et al. 2012; Vengoechea et al. 2013; Gao et al. 2013). All concluded that aberrant hypermethylation of subset of genes occurs in the early stage of meningiomas and highlighted the possibility of malignancy evaluation by assessing methylation status. However, a crucial methylation marker to predict an aggressive subtype in the early stage of meningiomas remains to be established, as existing data are still limited and often inconsistent. Our previous study has shown that a panel of 12 genes including ERCC1, hMLH1, ATM, CDKN2B (p15INK4B), p14ARF, CDKN2A (p16INK4A), RASSF1A, RUNX3, GATA6, NDRG2, PTEN, and RARβ might be useful in evaluation of glioma aggressiveness. In this regard, methylation index (MI) of these genes in individual patient, as well as RUNX3 methylation correlated with glioma WHO grade and higher MI, or aberrant methylation of RUNX3 indicated more aggressive tumors (Majchrzak-Celińska et al. 2015).
The aim of the present study was to evaluate the possible involvement of the hypermethylation of these genes in the malignant profile of benign and atypical meningiomas.
Materials and methods
Patients
The study included 44 patients who were referred to the Department and Clinic of Neurosurgery and Neurotraumatology, Poznan University of Medical Sciences, Poznań, Poland, between January 2010 and September 2013. Patients with a histologically confirmed diagnosis of meningioma at different grades of malignancy (I, I/II or II) and a treatment protocol that included surgical resection were enrolled in the study. Histological types of the tumors as well as tumor grades (according to the 2007 WHO classification criteria) were analyzed in the Laboratory of Neuropathology. Thirty-three patients were diagnosed with benign WHO grade I meningioma, two with grade I/II, and nine with atypical meningiomas (grade II). Women comprised 72.73 % (32/44) of all patients, and the average patients’ age was 56 years, ranging from 31–87. After resection, tumor samples were directly frozen and stored at −80 °C.
Control non-cancerous brain sample originated from a patient with brain hematoma, and blood sample for the analysis of white blood cells methylation was taken from healthy donor. All patients gave informed consent for the analyses to be undertaken, and the study protocol was approved by the Poznan University of Medical Sciences Clinical Research Ethics Committee (approval no. 505/12).
Isolation of DNA from tumor tissue
DNA was isolated using GeneMATRIX Tissue DNA Purification Kit [EurX, Gdańsk, Poland]. Briefly, 25 mg of tumor tissue was incubated with Lyse T Buffer, then RNase A and proteinase K, which was followed by incubation in 56 °C until the tissue was completely digested. Buffer Sol T and ethanol were then added to provide selective conditions for DNA binding during brief centrifugation, and then, DNA bound to minicolumn was washed twice to remove the traces of contaminants. DNA was eluted with Tris buffer, pH 8.5; its concentration and purity were measured using NanoDrop spectrophotometer. DNA was stored at −20 °C for further analysis.
Methylation-specific polymerase chain reaction (MSP)
Using MSP assay, the methylation status of ERCC1, hMLH1, ATM, CDKN2B (p15INK4B), p14ARF, CDKN2A (p16INK4A), RASSF1A, RUNX3, GATA6, NDRG2, PTEN, and RARβ was assessed. Bisulfite modification of 500 ng DNA, preceding MSP, was performed using the EZ DNA methylation kit [Zymo Research Corp., USA, D5002], following manufacturer’s protocol. Commercially available completely methylated control DNA [CpG Methylated HeLa Genomic DNA, New England Biolabs], completely unmethylated control DNA [EpiTect Control DNA, unmethylated, Qiagen], and water served as positive, negative, and blank controls of MSP assay, respectively. DNA samples derived from the white blood cells from healthy volunteers as well as a brain fragment from a patient with brain hematoma served as normal control samples for DNA methylation analysis.
MSP assay was carried out in MyCycler Thermal Cycler with Gradient [Bio-Rad] or T100 [Bio-Rad]. Primer sequences for all the analyzed genes with their corresponding annealing temperature are presented in Table 1. PCR reactions were carried out using either FastStart Taq DNA polymerase [Roche Diagnostics], TrueStart Hot Start Taq DNA polymerase [Fermentas], Perpetual Taq DNA polymerase [EurX], or My Taq HS DNA polymerase [Bioline].
The reaction conditions differed for each type of polymerase used, but generally after an initial step of heat denaturation at 95 °C for 1–4 min. (depending on the type of polymerase used), 35–38 cycles were carried out (95 °C for 15–30 s, appropriate annealing temperature for 30 s and 72 °C for 10–45 s), which were followed by final elongation at 72 °C for 5–7 min. The PCR products were separated by electrophoresis in 2 % agarose gel stained with ethidium bromide and visualized under UV illumination.
Pyrosequencing (PSQ)
Pyrosequencing (PSQ) of NDRG2 was performed on a PyroMark Vacuum Prep Workstation and Pyrosequencer PSQ™ 96 ID system, using PyroQ CpG™ software 1.0.9 (Biotage, Uppsala, Sweden), for quantification of the methylation level. Bisulfite-treated DNA samples were first amplified using FastStart Taq DNA Polymerase [Roche Diagnostics], with the forward primer: GGGCGGTGTGATTGATTC, and reverse, biotinylated primer: biotin-CCCACAATCTTCTCCCGTTCT (primers were designed using PyroQ CpG™ software 1.0.9). The 25 μl of reaction mixture contained 5 mM MgCl2, 10 × buffer, 0.2 mM dNTPs, 5 U/μl polymerase, 0.4 μM forward primer, 0.2 μl reverse primer, and water. The reaction conditions were as follows: 95 °C 4 min., 45 cycles of 95 °C for 30 s, 58 °C for 30 s, 72 °C for 45 s, followed by 72 °C for 7 min. The PCR product had 327 bp, and 2 μl of the product was visualized after ethidium bromide staining on 2 % agarose gel; the rest of the PCR product underwent PSQ. The analyzed region is presented in Fig. 1. The sequencing primer was as follows: GGGGGAAAGGGTTAGTA. The analyzed fragment spanned 5 CpG dinucleotides: T/CGGAGT/CGGGT/CGTTT/CGGTTGTTGT/CGTATAAAGGT, and the dispensation order was as follows: GTCGATGTCAGTCTGTCGTGTAGTCGTA.
As controls, completely methylated control DNA (CpG Methylated HeLa Genomic DNA, New England Biolabs), and completely unmethylated control DNA (EpiTect Control DNA, unmethylated, Qiagen), as well as DNA from normal, non-cancerous brain tissue were included in the assay. The reaction without any template DNA (blank) was also performed. Internal control for completion of the bisulfite treatment was included in the sequence. The cutoff value to distinguish between methylated and unmethylated samples was established at 11 %, based on the results obtained on control non-cancerous brain tissue, according to the previously described formula (Majchrzak-Celińska et al. 2015; Håvik et al. 2012).
Statistical analysis
Css Statistica, version 10, was used for statistical analysis. Fisher’s exact test was used to determine the correlations between methylation frequency and WHO grade of the tumors (Stat Exact program). Spearman’s rank correlation coefficient was used to measure the dependence between MI and WHO grade of the tumor, as well as patient’s age. Student’s t test was applied to determine the relevance of Pearson’s R coefficient. Chi-squared test and Pearson’s R coefficient were used to determine the correlation between methylation analysis results and patient’s age. Kruskal–Wallis test was used to determine the dependence between histological type of the tumor and MI, differences in methylation levels determined by PSQ between tumors of different WHO grade and the dependences between methylation levels, when analyzed with PSQ and patient’s age. Mann–Whitney U test was used to determine the statistical significance of methylation levels in each CpG site in terms of patient’s sex.
Results
MSP analysis
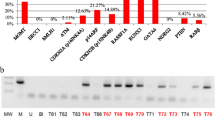
MSP analysis was applied to analyze the methylation status of 12 genes in 44 meningioma samples. The frequency of gene promoter methylation is presented in Fig. 2. The most frequently methylated gene was p14ARF—methylated in 31.82 % (14/44) of tumor samples, followed by RASSF1A methylated in 27.27 % (12/44) and CDKN2B (p15INK4B) which was methylated in 25.00 % (11/44) of cases. RUNX3 and GATA6 were found methylated in 15.91 % (7/44) and 13.64 % (6/44), respectively. Infrequent methylation was found for CDKN2A (p16INK4A) (9.09 %, 4/44), ATM (6.82 %, 3/44), and RARβ (2.38 %, 1/42) promoters. ERCC1, hMLH1, NDRG2, and PTEN were unmethylated in all samples analyzed (0 %, 0/44). The data illustrating the correlation of gene promoter methylation with WHO tumor grade are presented in Table 2. The median number of methylated genes was 1, and the highest MI was 0.33, indicating four methylated genes. RUNX3 methylation was statistically correlated with higher grade meningiomas (p = 0.03), and RASSF1A was more frequently methylated in men than women (p = 0.04).
NDRG2 pyrosequencing (PSQ)
Twenty-five meningioma samples were analyzed using PSQ. Sixty-eight percent of the samples (17/25) represented benign tumors (WHO grade I); however, one sample was classified as I/II grade, and seven were classified as grade II. Representative pyrogram is shown in Fig. 3. Totally, 84.00 % (21/25) of tumors showed a mean methylation signal above the level represented by normal, non-cancerous brain tissue. The highest methylation level was found for CpG4, whereas the lowest for CpG1. The average methylation of the five CpG sites analyzed in the methylated group were as follows: CpG1—8.17 %, CpG2—14.05 %, CpG3—17.14 %, CpG4—27.60 %, and CpG5—14.09 %. The mean methylation levels for tumor WHO grade I and I/II were 15.90 % (±5.53 %) and for grade II 13.71 % (±8.41 %) and thus could not differentiate between tumor WHO grades. The relationship between average NDRG2 methylation level in each CpG dinucleotide and WHO grades of analyzed meningiomas is presented in Fig. 4. Kruskal–Wallis test revealed that the average methylation level of all five CpG sites correlates with patient’s age—higher methylation levels were observed in older patients, as compared with younger ones (p = 0.03). According to Mann–Whitney’s U test, women had statistically higher methylation levels in CpG2 as compared to men (p = 0.01).
The comparison of NDRG2 promoter methylation profile obtained by MSP and PSQ revealed that 4 out of 25 samples gave concordant results when analyzed using MSP and PSQ, with the cutoff value determined to be 11 %. The high discordant rate was expected since two different fragments of the promoter were analyzed (the distance between them was 643 nt).
Discussion
Recent genome-scale DNA methylation analyses of different subtypes of meningiomas although provided novel insights into the molecular pathophysiology of malignant transformation did not indicate the definite markers of the development of these tumors (Kishida et al. 2012; Gao et al. 2013). Therefore, a crucial methylation marker to predict a greater malignant potential of meningiomas still remains to be established.
In our study, DNA methylation of a panel of genes was assessed in a group of patients with benign or atypical meningiomas. The panel included genes encoding DNA repair proteins (ERCC1, hMLH1, and ATM), cell cycle regulators (genes encoded in INK4B-ARF-INK4A locus, RASSF1A, and PTEN), and genes involved in cell differentiation and proliferation (RARβ, GATA6, NDRG2, and RUNX3). Our earlier study revealed that MI of these genes, i.e., the number of methylated genes divided by the number of all genes analyzed in the panel (Chen et al. 2011) correlated with glioma aggressiveness (Majchrzak-Celińska et al. 2015). The results of this study showed that such correlation does not occur in meningiomas. Although tumors of the same grade according to WHO criteria had differently methylated genes, the number of methylated genes for each individual patient was low, with the median of one and the maximum of four methylated genes out of 12. However, the analysis of individual genes methylation provided some interesting observations. The most important finding of our current study was the methylation of RUNX3, which was found in more than half of atypical meningiomas [55.56 %] and also in a subset of benign meningiomas [16.67 %]. More importantly, according to Fisher’s exact test, RUNX3 methylation correlated with the presence of more aggressive, atypical meningiomas (p = 0.03).
The Runt-related transcription factors (RUNX) are involved in developmental processes and, through multiple-interacting proteins, have been implicated in diverse signaling pathways and cellular processes (Chuang et al. 2013). The frequent inactivation of RUNX genes in cancer indicates crucial roles for RUNX in tumor suppression; however, they have also been characterized as oncogenes, depending on the context (Blyth et al. 2005). The role of RUNX3 methylation has been intensively studied in gastric cancers (Fan et al. 2011); however, data relating CNS tumors are very limited and apply only to small groups of patients (Avci et al. 2014). Thus, although the results of our present study require confirmation on larger patient cohorts, they suggest that RUNX3 methylation might be considered as the promising candidate biomarker for the assessment of meningioma aggressivenes.
Recently, significant differences in the expression of NDRG2 gene were reported between primary and recurrent meningioma groups, and benign and atypical meningiomas (Skiriute et al. 2011). Earlier, the downregulation of this gene was described in a subset of lower-grade meningiomas, including atypical meningiomas with clinically aggressive behavior, which was associated with hypermethylation of the NDRG2 promoter (Lusis et al. 2005). In this study, we analyzed NDRG2 promoter methylation using PSQ technique, because the sequence, reported to be a target of aberrant methylation in colon cancer (Piepoli et al. 2009) and used in our MSP analysis, was found unmethylated in all meningioma samples analyzed. The analysis of five CpG sites in NDRG2 promoter revealed methylation in a large subset of meningiomas. Eighty-four percent of tumor samples showed a mean methylation signal above the level found in normal, non-cancerous brain tissue. However, the mean methylation levels of five CpGs were below 20 % for all meningioma grades and whether this mild methylation has any significant impact on mRNA levels remains to be determined. However, higher methylation levels in CpG2 were observed in women than men, and the average methylation level of all five CpG sites correlated with patient’s age. Thus, the results of our study confirm that mild NDRG2 methylation is a common event in meningioma formation, but clinical application of this observation seems to be limited. Moreover, PSQ analysis of a sequence within 5′UTR was proven to be a reliable method for its analysis.
Methylation of RASSF1A promoter is considered as an early and frequent event in tumorigenesis, and there are attempts to apply RASSF1A methylation as a diagnostic marker in cancer screening (Richter et al. 2009). The results of our study revealed that RASSF1A is methylated in 27.27 % of meningioma samples, and the frequency of gene promoter methylation is increasing with the tumor grade (25.00 % of tumors grade I and I/II, and 55.56 % of tumors grade II). Thus, we confirm the earlier findings of Nakane et al. (2007), which showed RASSF1A promoter methylation in 18.2, 63.6, and 42.9 % of the grade I, II, and III of meninigiomas, respectively. Overall, these results suggest that RASSF1A methylation is likely to play an important role in both meningioma development and progression. However, because this gene is frequently methylated in other malignancies (Richter et al. 2009), its role as meningioma biomarker is limited and should be considered only in a panel of other frequently methylated genes.
The panel of genes whose promoter methylation was analyzed in this study included also other commonly hypermethylated ones, such as those located in the INK4B-ARF-INK4A locus. Inactivation of the G1/S-phase cell cycle checkpoint was reported to be an important aberration in anaplastic meningiomas (Boström et al. 2001), and it was suggested that deregulations of p14-MDM2-p53 pathway may contribute to the malignant progression of those tumors (Amatya et al. 2004). The results of our study pointed to p14ARF gene as the most frequently methylated one in INK4B-ARF-INK4A locus (31.82 % of all samples), followed by p15INK4B (CDKN2B) (25.00 %) and p16INK4A (CDKN2A) (9.09 %). Methylation of these genes characterized both benign as well as atypical meningiomas. These findings indicate that INK4B-ARF-INK4A locus is a target of aberrant methylation in meningiomas and that at least in some of those tumors the loss of cell cycle control is due to epigenetic silencing of p14ARF, p15INK4B, and/or p16INK4A genes. According to the two-hit model of Knudson, DNA methylation can be a second hit inactivating those tumor suppressors, following genetic mutations which are also often present in meningiomas in this locus (Boström et al. 2001; Knudson 1996). Similar findings concerning DNA methylation in meningiomas were recently published by Aydemir et al. (2012), who found p16INK4A to be methylated in 8.3 % [3/36] of cases and Liu et al. (2005) reporting methylation of this gene in 10 % of samples. Amatya et al. (2004) found methylation of p14ARF gene in 5 of 58 cases of benign meningiomas (8.6 %), 2 of 10 cases of atypical meningiomas (20 %), and 2 of 4 cases of anaplastic meningiomas (50 %). However, the utility of INK4B-ARF-INK4A locus methylation in terms of clinically useful biomarker seems limited. Linsler et al. (2014) reported that methylation of p16INK4A is not associated with recurrence, higher grade, or chromosomal aberrations observed in meningiomas. What is more neither age nor sex had significant influence on tumor recurrence, with regard to the p16INK4A methylation status (Linsler et al. 2014).
Recently, GATA6 gene was found to be frequently methylated in glioma patients (Skiriute et al. 2012; Cecener et al. 2012; Martinez et al. 2009). Moreover, significant association of this gene methylation with unfavorable patient survival was found (Martinez et al. 2009). In meningiomas, to our best knowledge, the methylation of this gene has not been analyzed. Our results indicate that similarly to gliomas, GATA6 promoter is also methylated in meningiomas, however, with significantly lower frequency (13.64 vs 30–68 % in glioma). Thus, GATA6 promoter methylation in meningioma may play certain role in menigioma development, although cannot be considered as a biomarker candidate.
The other genes analyzed in this study, namely ATM and RARβ, were found to be methylated in a marginal number of patients (ATM—6.82 %, RARβ—2.38 %), while ERCC1, hMLH1, and PTEN were not methylated at all. The low frequency or lack of methylation of those genes suggests that their promoter methylation does not play a major role in the development of meningioma.
In summary, this study provided evidence that p14ARF, RASSF1A, p15INK4B (CDKN2B), RUNX3, GATA6, p16INK4A (CDKN2A), NDRG2 and to lesser extent ATM and RARβ promoter methylation are associated with the development of meningiomas. RUNX3 methylation correlates with meningioma WHO grade and, therefore, can be used as a potential indicator of tumor aggressiveness. Despite the methylation of NDRG2 in the majority of meningiomas, it cannot be considered as a diagnostic biomarker because its methylation level is only slightly elevated in comparison to non-cancerous brain tissue. Overall, the results of this study confirm that DNA methylation plays an important role in the pathogenesis of meningiomas. Further investigations, particularly concerning RUNX3 methylation, are required in order to assess clinically useful potential of the genes analyzed in this study.
References
Amatya VJ, Takeshima Y, Inai K (2004) Methylation of p14(ARF) gene in meningiomas and its correlation to the p53 expression and mutation. Mod Pathol 17:705–710
Avci CB, Dodurga Y, Susluer SY, Sıgva ZO, Yucebas M, Caglar HO, Akalin T, Dalbasti T, Oktar N, Gunduz C (2014) Promoter hypermethylation-mediated down-regulation of RUNX3 gene in human brain tumors. Ir J Med Sci 183:259–264
Aydemir F, Yurtcu E, Balci TB, Sahin FI, Gulsen S, Altinors N (2012) Identification of promoter region methylation patterns of MGMT, CDKN2A, GSTP1, and THBS1 genes in intracranial meningioma patients. Genet Test Mol Biomarkers 16:335–340
Barski D, Wolter M, Reifenberger G, Riemenschneider MJ (2010) Hypermethylation and transcriptional downregulation of the TIMP3 gene is associated with allelic loss on 22q12.3 and malignancy in meningiomas. Brain Pathol 20:623–631
Blyth K, Cameron ER, Neil JC (2005) The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer 5:376–387
Boström J, Meyer-Puttlitz B, Wolter M, Blaschke B, Weber RG, Lichter P, Ichimura K, Collins VP, Reifenberger G (2001) Alterations of the tumor suppressor genes CDKN2A (p16(INK4a)), p14(ARF), CDKN2B (p15(INK4b)), and CDKN2C (p18(INK4c)) in atypical and anaplastic meningiomas. Am J Pathol 159:661–669
Cahill KS, Claus EB (2011) Treatment and survival of patients with nonmalignant intracranial meningioma: results from the surveillance, epidemiology, and end results program of the National Cancer Institute. Clinical article. J Neurosurg 115:259–267
Cecener G, Tunca B, Egeli U, Bekar A, Tezcan G, Erturk E, Bayram N, Tolunay S (2012) The promoter hypermethylation status of GATA6, MGMT, and FHIT in glioblastoma. Cell Mol Neurobiol 32:237–244
Chen PC, Tsai MH, Yip SK, Jou YC, Ng CF, Chen Y, Wang X, Huang W, Tung CL, Chen GC, Huang MM, Tong JH, Song EJ, Chang DC, Hsu CD, To KF, Shen CH, Chan MW (2011) Distinct DNA methylation epigenotypes in bladder cancer from different Chinese sub-populations and its implication in cancer detection using voided urine. BMC Med Genomics. doi:10.1186/1755-8794-4-45
Chuang LS, Ito K, Ito Y (2013) RUNX family: regulation and diversification of roles through interacting proteins. Int J Cancer 132:1260–1271
Dolecek TA, Propp JM, Stroup NE, Kruchko C (2012) CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro Oncol. doi:10.1093/neuonc/nos218
Fan XY, Hu XL, Han TM, Wang NN, Zhu YM, Hu W, Ma ZH, Zhang CJ, Xu X, Ye ZY, Han CM, Pan WS (2011) Association between RUNX3 promoter methylation and gastric cancer: a meta-analysis. BMC Gastroenterol. doi:10.1186/1471-230X-11-92
Gao F, Shi L, Russin J, Zeng L, Chang X, He S, Chen TC, Giannotta SL, Weisenberger DJ, Zada G, Mack WJ, Wang K (2013) DNA methylation in the malignant transformation of meningiomas. PLoS One. doi:10.1371/journal.pone.0054114
Håvik AB, Brandal P, Honne H, Dahlback HS, Scheie D, Hektoen M, Meling TR, Helseth E, Heim S, Lothe RA, Lind GE (2012) MGMT promoter methylation in gliomas-assessment by pyrosequencing and quantitative methylation-specific PCR. J Transl Med 10:36
Kishida Y, Natsume A, Kondo Y, Takeuchi I, An B, Okamoto Y, Shinjo K, Saito K, Ando H, Ohka F, Sekido Y, Wakabayashi T (2012) Epigenetic subclassification of meningiomas based on genome-wide DNA methylation analyses. Carcinogenesis 33:436–441
Knudson AG (1996) Hereditary cancer: two hits revisited. J Cancer Res Clin Oncol 122:135–140
Lamszus K (2004) Meningioma pathology, genetics, and biology. J Neuropathol Exp Neurol 63:275–286
Linsler S, Kraemer D, Driess C, Oertel J, Kammers K, Rahnenführer J, Ketter R, Urbschat S (2014) Molecular biological determinations of meningioma progression and recurrence. PLoS One. doi:10.1371/journal.pone.0094987
Liu Y, Pang JC, Dong S, Mao B, Poon WS, Ng HK (2005) Aberrant CpG island hypermethylation profile is associated with atypical and anaplastic meningiomas. Hum Pathol 36:416–425
Lusis EA, Watson MA, Chicoine MR, Lyman M, Roerig P, Reifenberger G, Gutmann DH, Perry A (2005) Integrative genomic analysis identifies NDRG2 as a candidate tumor suppressor gene frequently inactivated in clinically aggressive meningioma. Cancer Res 65:7121–7126
Majchrzak-Celińska A, Paluszczak J, Szalata M, Barciszewska AM, Nowak S, Kleszcz R, Sherba A, Baer-Dubowska W (2015) The methylation of a panel of genes differentiates low-grade from high-grade gliomas. Tumour Biol. doi:10.1007/s13277-014-3025-3
Martinez R, Martin-Subero JI, Rohde V, Kirsch M, Alaminos M, Fernandez AF, Ropero S, Schackert G, Esteller M (2009) A microarray-based DNA methylation study of glioblastoma multiforme. Epigenetics 4:255–264
Nakane Y, Natsume A, Wakabayashi T, Oi S, Ito M, Inao S, Saito K, Yoshida J (2007) Malignant transformation-related genes in meningiomas: allelic loss on 1p36 and methylation status of p73 and RASSF1A. J Neurosurg 107:398–404
Paluszczak J, Baer-Dubowska W (2006) Epigenetic diagnostics of cancer—the application of DNA methylation markers. J Appl Genet 47:365–375
Piepoli A, Cotugno R, Merla G, Gentile A, Augello B, Quitadamo M, Merla A, Panza A, Carella M, Maglietta R, D’Addabbo A, Ancona N, Fusilli S, Perri F, Andriulli A (2009) Promoter methylation correlates with reduced NDRG2 expression in advanced colon tumour. BMC Med Genomics. doi:10.1186/1755-8794-2-11
Richter AM, Pfeifer GP, Dammann RH (2009) The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim Biophys Acta 1796:114–128
Skiriute D, Tamasauskas S, Asmoniene V, Saferis V, Skauminas K, Deltuva V, Tamasauskas A (2011) Tumor grade-related NDRG2 gene expression in primary and recurrent intracranial meningiomas. J Neurooncol 102:89–94. doi:10.1007/s11060-010-0291-9
Skiriute D, Vaitkiene P, Saferis V, Asmoniene V, Skauminas K, Deltuva VP, Tamasauskas A (2012) MGMT, GATA6, CD81, DR4, and CASP8 gene promoter methylation in glioblastoma. BMC Cancer. doi:10.1186/1471-2407-12-218
Vengoechea J, Sloan AE, Chen Y, Guan X, Ostrom QT, Kerstetter A, Capella D, Cohen ML, Wolinsky Y, Devine K, Selman W, Barnett GH, Warnick RE, McPherson C, Chiocca EA, Elder JB, Barnholtz-Sloan JS (2013) Methylation markers of malignant potential in meningiomas. J Neurosurg 119:899–906
Acknowledgments
This study was supported by National Science Centre of Poland, Grant No. NN405 683 240.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Majchrzak-Celińska, A., Paluszczak, J., Szalata, M. et al. DNA methylation analysis of benign and atypical meningiomas: correlation between RUNX3 methylation and WHO grade. J Cancer Res Clin Oncol 141, 1593–1601 (2015). https://doi.org/10.1007/s00432-015-1930-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-015-1930-5