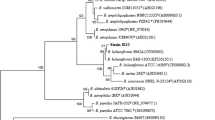
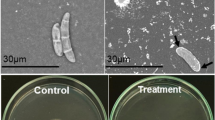
Abstract
Microbes have many mechanisms to exert their inhibitory activity against target pests. One such mechanism involves the production and secretion of hydrolytic enzymes, such as chitinases, which are produced naturally by plants in response to attack by insect herbivores and phytopathogens and have been sought as an additional factor to enhance pest management. Thus, our main aim was to screen the diverse actinomycete community associated with the integument of Acromyrmex subterraneus brunneus for a chitinase-producing strain and to characterize its chitinases. We identified isolate ENT-21—a Streptomyces sp.—as a chitinase-producer and our data indicate that this isolate produces a chitinolytic complex that contains a chitinase and a high-molecular-weight β-N-acetylglucosaminidase (>100 kDa) when cultured in Chitin-Czapek broth. The presence of chitinases in the genome of this isolate was checked by diagnostic PCR, and two chitinase genes belonging to family 18 group A and family 19 were verified. The chitinolytic activity of the crude extract was observed at pH values ranging from 3.8 to 11.0, with the highest chitinase activities recorded at pH 9.0 and 9.5, whereas optimum β-N-acetylglucosaminidase activity was observed over a narrow pH range, between pH 4.7 and 5.1. We describe some biochemical and molecular properties of the chitinase and β-N-acetylglucosaminidase produced by ENT-21, and discuss the potential for exploitation of these enzymes for pest control.





Similar content being viewed by others
References
Arakane Y, Muthukrishnan S (2010) Insect chitinase and chitinase-like proteins. Cell Mol Life Sci 67:201–216
Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Bhattacharya D, Nagpure A, Gupta RK (2007) Bacterial chitinases: Properties and potential. Crit Rev Biotechnol 27:21–28
Bradford MM (1976) A rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal Biochem 72:248–254
Chandrasekaran R, Revathi K, Nisha S, Kirubakaran SA, Sathish-Narayanan S, Senthil-Nathan S (2012) Physiological effect of chitinase purified from Bacillus subtilis against the tobacco cutworm Spodoptera litura Fab. Pestic Biochem Physiol 104:65–71
Chater KF, Biró S, Lee KJ, Palmer T, Schrempf H (2010) The complex extracellular biology of Streptomyces. FEMS Microbiol Rev 34:171–198
Currie CR, Poulsen M, Mendenhall J, Boomsma JJ, Billen J (2006) Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science 6:81–83
Dahiya N, Tewari R, Hoondal GS (2006) Biotechnological aspects of chitinolytic enzymes: a review. Appl Microbiol Biotechnol 71:773–782
Das SN, Neeraja C, Sarma PVSRN, Prakash JM, Purushotham P, Kaur M, Dutta S, Podile AR (2012) Microbial chitinases for chitin waste management. In: Satyanarayana T, Johri BN, Prakash A (eds) Microorganisms in environmental management. Springer, Dordrecht, pp 135–150
Dawson RMC, Elliott DC, Elliott WH, Jones KM (eds) (1989) Data for biochemical research. Oxford University Press, NewYork
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Filho BPD, Lemos FJA, Secundino NFC, Pascoa V, Pereira ST, Pimenta PFP (2002) Presence of chitinase and β-N-acetylglucosaminidase in the Aedes aegypti: a chitinolytic system involving peritrophic matrix formation and degradation. Insect Biochem Mol Biol 32:1723–1729
Fitch WM (1971) Toward defining the course of evolution: minimum change for specific tree topology. Syst Biol 20:406–416
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Guo YP, Zheng W, Rong XY, Huang Y (2008) A multilocus phylogeny of the Streptomyces griseus 16S rRNA gene clade: use of multilocus sequence analysis for streptomycete systematics. Int J Syst Evol Microbiol 58:149–159
Hahn M, Hennig M, Schlesier B, Höhne W (2000) Structure of jack bean chitinase. Acta Crystallogr D 56:1096–1099
Herrera-Estrella A, Chet I (1999) Chitinases in biological control. Chitin Chitinases 87:171–184
Horsch M, Mayer C, Sennhauser U, Rast DM (1997) β-N-acetylhexosaminidase: a target for the design of antifungal agents. Pharmacol Ther 76:187–218
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism, vol 3. Academic, New York, pp 21–123
Kawase T, Saito A, Sato T, Kanai R, Fujii T, Nikaidou N, Miyashita N, Watanabe T (2004) Distribution and phylogenetic analysis of famly 19 chitinases in Actinobacteria. Appl Environ Microbiol 70(2):1135–1144
Kawase T, Yokokawa S, Saito A, Fujii T, Nikaidou N, Miyashita K, Watanabe T (2006) Comparison of enzymatic and antifungal properties between family 18 and 19 chitinases from S. coelicolor A3(2). Biosci Biotechnol Biochem 70(4):988–998
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kramer KJ, Muthukrishnan S (1998) Insect chitinases: molecular biology and potential use as biopesticides. Insect Biochem Mol Biol 27:887–900
Labeda DP, Goodfellow M, Brown R, Ward AC, Lanoot B, Vanncanneyt M, Swings J, Kim SB, Liu Z, Chun J, Tamura T, Oguchi A, Kikuchi T, Kikuchi H, Nishii T, Tsuji K, Yamaguchi Y, Tase A, Takahashi M, Sakane T, Suzuki KI, Hatano K (2012) Phylogenetic study of the species within the family Streptomycetaceae. Antonie Van Leeuwenhoek 101:73–104
Lehane MJ (1997) Peritrophic matrix structure and function. Annu Rev Entomol 42:525–550
Lehane MJ, Billingsley PF (1996) Biology of the insect midgut. Chapman & Hall, London
Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39(D):225–229
Marco JL, Valadares-Inglis MC, Felix CR (2004) Purification and characterization of an N-acetylglucosaminidase produced by a Trichoderma harzianum strain which controls Crinipellis perniciosa. Appl Microbiol Biotechnol 64:70–75
Martínez CP, Echeverri C, Florez JC, Gaitan AL, Góngora CE (2012) In vitro production of two chitinolytic proteins with an inhibiting effect on the insect coffee berry borer, Hypothenemus hampei (Ferrari) (Coleoptera: Curculionidae) and the fungus Hemileia vastatrix the most limiting pests of coffee crops. AMB Express 2:22 [http://www.amb-express.com/content/2/1/22]
Mizuno R, Fukamizo T, Sugiyama S, Nishizawa Y, Kezuka Y, Nonaka T, Suzuki K, Watanabe T (2008) Role of the loop structure of the catalytic domain in rice class I chitinase. J Biochem 143:487–495
Nagpure A, Gupta RK (2013) Purification and characterization of an extracellular chitinase from antagonistic Streptomyces violaceusniger. J Basic Microbiol 63:871–877
Nawani NN, Kapadnis BP (2005) Optimization of chitinase production using statistics based experimental designs. Process Biochem 40:651–660
Oppenheim AB, Chet I (1992) Cloned chitinases in fungal plant-pathogen control strategies. Trends Biotechnol 10:392–394
Ramı́rez MG, Avelizapa LIR, Avelizapa NGR, Camarillo RC (2004) Colloidal chitin stained with Remazol Brilliant Blue R®, a useful substrate to select chitinolytic microorganisms and to evaluate chitinases. J Microbiol Methods 56(2):213–219
Rong XY, Huang Y (2010) Taxonomic evaluation of the Streptomyces griseus clade using multilocus sequence analysis and DNA–DNA hybridization, with proposal to combine 29 species and three subspecies as 11 genomic species. Int J Syst Evol Microbiol 60:696–703
Rong XY, Huang Y (2012) Taxonomic evaluation of the Streptomyces hygroscopicus clade using multilocus sequence analysis and DNA–DNA hybridization, validating the MLSA scheme for systematics of the whole genus. Syst Appl Microbiol 35(1):7–18
Rong XY, Guo YP, Huang Y (2009) Proposal to reclassify the Streptomyces albidoflavus clade on the basis of multilocus sequence analysis and DNA-DNA hybridization, and taxonomic elucidation of Streptomyces griseus subsp. solvifaciens. Syst Appl Microbiol 32:314–322
Rosselló-Mora R, Amann R (2001) The species concept for prokaryotes. FEMS Microbiol Rev 25(1):39–67
Saitou N, Nei M (1987) The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:404–425
Sambrook J, Russell DW (2001) Molecular cloning—a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Shahidi F, Kamil YVAJ (2001) Enzymes from fish and aquatic invertebrates and their application in the food industry. Trends Food Sci Technol 12:435–464
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Evol Microbiol 16:313–340
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Terra WR, Ferreira C (2005) Biochemistry of digestion. In: Gilbert LI, Iatrov K, Gill S (eds) Comprehensive Molecular Insect Science 4:171–224. Elsevier, Oxford
Terra WR, Ferreira C, Bianchi AGD (1979) Distribution of digestive enzymes among the endoperitrophic and ectoperitrophic spaces and midgut cells of Rhynchosciara and its physiological significance. J Insect Physiol 25:487–494
Tharanathan RN, Kittur FS (2003) Chitin—the undisputed biomolecule of great potential. Crit Rev Food Sci 43:61–87
Usui T, Matsui H, Isobe K (1990) Enzymic-synthesis of useful chito-oligosaccharides utilizing transglycosylation by chitinolytic enzymes in a buffer containing ammonium-sulfate. Carbohydr Res 203:65–77
Wang F, Xiao X, Saito A, Schrempf H (2002) Streptomyces olivaceoviridis possesses a phosphotransferase system that mediates specific, phosphoenolpyruvate-dependent uptake of N-acetylglucosamine. Mol Genet Genomics 268:344–351
Watanabe T, Kanai R, Kawase T, Tanabe T, Mitsutomi M, Sakuda S, Miyashita K (1999) Family 19 chitinases of Streptomyces species: characterization and distribution. Microbiology 145:3353–3363
Williamson N, Brian P, Wellington EMH (2000) Molecular detection of bacterial and streptomycete chitinases in the environment. Antonie Van Leeuwenhoek 78:315–321
Wu Y, Liu F, Liu Y, Zhang Z, Zhou T, Liu X, Shen Q, Shen B (2011) Identification of chitinases Is-chiA and Is-chiB from Isoptericola jiangsuensis CLG and their characterization. Appl Microbiol Biotechnol 89:705–713
Zucchi TD, Almeida LG, Dossi FCA, Cônsoli FL (2010) Secondary metabolites produced by Propionicimonas sp. (ENT-18) induce histological abnormalities in the sclerotia of Sclerotinia sclerotiorum. BioControl 55:811–819
Zucchi TD, Guidolin AS, Consoli FL (2011a) Isolation and characterization of actinobacteria ectosymbionts from Acromyrmex subterraneus brunneus (Hymenoptera, Formicidae). Microbiol Res 166:68–76
Zucchi TD, Rossi GD, Cônsoli FL (2011b) Characterization of a β-amylase from Propionicimonas sp. ENT-18 ectosymbiont of Acromyrmex subterraneus brunneus. Ann Microbiol 61:985–990
Zucchi TD, Almeida LG, Moraes LAB, Cônsoli FL (2014) Albocycline, the main bioactive compound from Propionicimonas sp. ENT-18 against Sclerotinia sclerotiorum. Ind Crops Prod 52C:264–268
Acknowledgments
The authors appreciate the financial support provided by FAPESP to G.D.R. (PhD fellowship—process 2012/50021-1), T.D.Z. (PostDoctoral fellowship—processes 2007/58712-5 and 2011/14333-6), A.S.G. (PhD fellowship—process 2012/04287-0) and A.P. (Masters fellowship—process 2010/13675-8). F.L.C. also acknowledges FAPESP for funding this research (grants—processes 2007/59019-1; 2011/50877-0).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Rossi, G.D., Zucchi, T.D., Guidolin, A.S. et al. Chitin-degrading enzymes from an actinomycete ectosymbiont of Acromyrmex subterraneus brunneus (Hymenoptera: Formicidae). Ann Microbiol 65, 565–574 (2015). https://doi.org/10.1007/s13213-014-0892-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-014-0892-1