Abstract
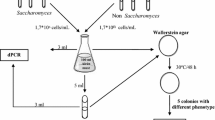
The aim of this study was to investigate the yeast and fungal microflora diversity in wine-related samples of the cultivars Veltlín zelený (Grüner Veltliner) and Frankovka modrá (Blaufränkisch) produced in the Small Carpathian region (Slovakia) using different eukaryotic identification approaches. For phenotypic identification we performed auxotrophy profiling using the API 20C AUX kit (bioMérieux, Marcy l'Étoile, France) and culture on selective lysine medium. Genotypic identification was done by real-time PCR assays specific for different wine yeasts and by sequencing of the internal transcribed spaced (ITS) region (ITS1–5.8S rRNA–ITS2 region). An effective culture-independent approach was also employed in the form of analysis of the ITS2 region using fluorescence–ITS PCR (f-ITS PCR), cloning and sequencing. The ITS sequencing results were concordant with the results of the selective culture and real-time PCR assay, but for several strains they did not agree with the results of auxotrophy profiling. In the culture-independent approach, sequences of clones included 100 % matches to Hanseniaspora uvarum, Candida zemplinina, Metschnikowia pulcherrima, Aureobasidium pullulans, Saccharomyces cerevisiae and Issatchenkia terricola, as well as two novel sequences with a 5-bp difference to Sporobolomyces marcillae. The data from this study, and particularly the data from the culture-independent investigation, contribute to the picture of the wine microbial community of two cultivars important in wine production in Slovakia and Central Europe.


Similar content being viewed by others
References
Allmann M, Candrian U, Höfelein C, Lüthy J (1993) Polymerase chain reaction (PCR). A possible alternative to immunochemical methods assuring safety and quality of food. Z Lebensm Unters Forsch 196:248–251
Arias CR, Burns JK, Friedrich LM, Goodrich RM, Parish ME (2002) Yeast species associated with orange juice: evaluation of different identification methods. Appl Environ Microbiol 68:1955–1961
Blondin B, Vezinhet F (1988) Identification de souches de levures oenologiques par leurs caryotypes obtenus en électrophorèse en champ pulsé. Rev Fr Oenol 28:7–11
Brežná B, Ženišová K, Chovanová K, Chebeňová V, Kraková L, Kuchta T, Pangallo D (2010) Evaluation of fungal and yeast diversity in Slovakian wine-related microbial communities. Antonie Van Leeuwenhoek 98:519–529
Chovanová K, Kraková L, Ženišová K, Turcovská V, Brežná B, Kuchta T, Pangallo D (2011) Selection and identification of autochthonous yeasts in Slovakian wine samples using a rapid and reliable three-step approach. Lett Appl Microbiol 53:231–237
Cocolin L, Campolongo S, Alessandria V, Dolci P, Rantsiou K (2011) Culture independent analyses and wine fermentation: an overview of achievements 10 years after first application. Ann Microbiol 61:17–23
Degré R, Thomas DY, Ash J, Mailhiot K, Monn A, Dubord C (1989) Wine yeasts strain identification. Am J Enol Vitic 40:309–315
Di Maro E, Ercolini D, Coppola S (2007) Yeast dynamics during spontaneous wine fermentation of the Catalanesca grape. Int J Food Microbiol 117201–210
Fechner LC, Vincent-Hubert F, Gaubert P, Bouchez T, Gourlay-Francé C, Tusseau-Vuillemin MH (2010) Combined eukaryotic and bacterial community fingerprinting of natural freshwater biofilms using automated ribosomal intergenic spacer analysis. FEMS Microbiol Ecol 74:542–553
Fernández-González M, Briones AI (2013) Study of yeast populations and their enological properties in Guijoso Appellation of Origin (Spain). Ann Microbiol 63:371–379
Fleet GH, Prakitchaiwattana CJ, Beh AL, Heard GM (2002) The yeast ecology of wine grapes. In: Ciani M (ed) Biodiversity and biotechnology of wine yeasts. Research Signpost, Kerala, pp 1–17
Francesca N, Chiurazzi M, Romano R, Aponte M, Settanni L, Moschetti G (2010) Indigenous yeast communities in the environment of “Rovello bianco” grape variety and their use in commercial white wine fermentation. World J Microbiol Biotechnol 26:337–351
Heard GM, Fleet GH (1986) Evaluation of selective media for enumeration of yeasts during wine fermentation. J Appl Microbiol 60:477–481
Ivey ML, Phister TG (2011) Detection and identification of microorganisms in wine: a review of molecular techniques. J Ind Microbiol Biotechnol 38:1619–1634
Kraková L, Chovanová K, Ženišová K, Turcovská V, Brežná B, Kuchta T, Pangallo D (2012) Yeast diversity investigation of wine-related samples from two different Slovakian wine-producing areas through a multistep procedure. LWT Food Sci Technol 46:406–411
Lopandic K, Tiefenbrunner W, Gangl H, Mandl K, Berger S, Leitner G, Abd-Ellah GA, Querol A, Gardner RC, Sterflinger K, Prillinger H (2008) Molecular profiling of yeasts isolated during spontaneous fermentations of Austrian wines. FEMS Yeast Res 8:1063–1075
Mannazzu I, Clementi F, Ciani M (2002) Strategies and criteria for the isolation and selection of autochtonous starters. In: Ciani M (ed) Biodiversity and biotechnology of wine yeasts. Research Signpost, Kerala, pp 19–33
Mills DA, Johannsen EA, Cocolin L (2002) Yeast diversity and persistence in botrytis-affected wine fermentations. Appl Environ Microbiol 68:4884–4893
Ness F, Lavallée F, Dubourdieu D, Aigle M, Dulau L (1993) Identification of yeast strains using the polymerase chain reaction. J Sci Food Agric 62:89–94
Nisiotou AA, Gibson GR (2005) Isolation of culturable yeasts from market wines and evaluation of the 5.8S-ITS rDNA sequence analysis for identification purposes. Lett Appl Microbiol 41:454–463
Querol A, Barrio E, Huerta T, Ramón D (1992a) Molecular monitoring of wine fermentations conducted by active dry yeast strains. Appl Environ Microbiol 58:2948–2953
Querol A, Barrio E, Ramón D (1992b) A comparative study of different methods of yeast strain characterization. Syst Appl Microbiol 15:439–446
Šuranská H, Vránová D, Omelková J, Vadkertiová R (2012) Monitoring of yeast population isolated during spontaneous fermentation of Moravian wine. Chem Zvesti 66:861–868
Taqarort N, Echairi A, Chaussod R, Nouaim R, Boubaker H, Benaoumar AA, Boudyach E (2008) Screening and identification of epiphytic yeasts with potential for biological control of green mold of citrus fruits. World J Microbiol Biotechnol 24:3031–3038
Tofalo R, Schirone M, Telera GC, Manetta AC, Corsetti A, Suzzi G (2011) Influence of organic viticulture on non-Saccharomyces wine yeast populations. Ann Microbiol 61:57–66
Torzilli AP, Sikaroodi M, Chalkley D, Gillevet PM (2006) A comparison of fungal communities from four salt marsh plants using automated ribosomal intergenic spacer analysis (ARISA). Mycologia 98:690–698
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, New York, pp 315–321
Zott K, Claisse O, Lucas P, Coulon J, Lonvaud-Funel A, Masneuf-Pomarede I (2010) Characterization of the yeast ecosystem in grape must and wine using real-time PCR. Food Microbiol 27:559–567
Acknowledgments
This study was supported by the Slovak Research and Development Agency under the contract No. APVV-0219-07.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ženišová, K., Chovanová, K., Chebeňová-Turcovská, V. et al. Mapping of wine yeast and fungal diversity in the Small Carpathian wine-producing region (Slovakia): evaluation of phenotypic, genotypic and culture-independent approaches. Ann Microbiol 64, 1819–1828 (2014). https://doi.org/10.1007/s13213-014-0827-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-014-0827-x