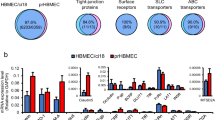
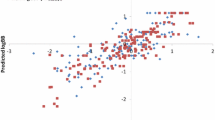
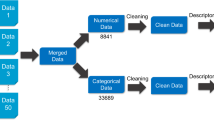
Abstract
An ideal neurotherapeutics agent for central nervous system (CNS) molecular targets should cross Blood Brain Barrier (BBB) whereas peripherally acting agent should not cross BBB to avoid CNS related side effects. Hence prediction of BBB permeability index has been proved to be an efficient tool for drug discovery and development. Various experimental and computational approaches are being used in recent past for the prediction of BBB permeability and they have been successful up to some extent. However the accuracy and authenticity of these methods have been under question. The current review article attempts to answer a vital question that is, can we predict BBB permeability using computational approaches.
Similar content being viewed by others
References
Abraham, M.H., Chadha, H.S., Mitchell, R.C. 1994. Hydrogen bonding. 33. Factors that influence the distribution of solutes between blood and brain. J Pharm Sci 83, 1257–1268.
Abraham, M.H., Chadha, H.S., Mitchell, R.C. 1995. Hydrogen-bonding. Part 36. Determination of blood brain distribution using octanol-water partition coefficients. Drug Des Discov 13, 123–131.
Brewster, M.E., Pop, E., Huang, M.J., Bodor, N. 1996. AM1-based model system for estimation of brain/blood concentration ratios. Int J Quantum Chem 60, 51–63.
Clark, D.E. 1999. Rapid calculation of polar molecular surface area and its application to the prediction of transport phenomena. 2. Prediction of blood-brain barrier penetration. J Pharm Sci 88, 815–821.
Crivori, P., Cruciani, G., Carrupt, P.A., Testa, B. 2000. Predicting blood-brain barrier permeation from three-dimensional molecular structure. J Med Chem 43, 2204–2216.
Doniger, S., Hofmann, T., Yeh, J. 2002. Predicting CNS permeability of drug molecules: Comparison of neural network and support vector machine algorithms. J Comput Biol 9, 849–864.
Dureja, H., Madan, A.K. 2007. Validation of topochemical models for the prediction of permeability through the blood-brain barrier. Acta Pharm 57, 451–467.
El-Deeb, I.M., Bayoumi, S.M., El-Sherbeny, M.A., Abdel-Aziz, A.A.M. 2010. Synthesis and antitumor evaluation of novel cyclic arylsulfonylureas: ADMET and pharmacophore prediction. Eur J of Med Chem 45, 2516–2530.
Fan, Y., Unwalla, R., Denny, R.A., Di, L., Kerns, E.H., Diller, D.J., Humblet, C. 2010. Insights for predicting blood-brain barrier penetration of CNS targeted molecules using QSPR approaches. J Chem Inf Model 50, 1123–1133.
Feher, M., Sourial, E., Schmidt, J.M. 2000. A simple model for the prediction of blood-brain partitioning. Int J Pharm 201, 239–247.
Garg, P., Verma, J., Roy, N. 2008. In silico modeling for blood brain barrier permeability predictions. In: Drug Absorption Studies, Volume VII, Springer, US, 510–544.
Geldenhuys, W.J., Manda, V.K., Mittapalli, R.K., van der Schyf, C.J., Crooks, P.A., Dwoskin, L.P., Allen, D.D., Lockman, P.R. 2010. Predictive screening model for potential vector-mediated transport ofcationic substrates at the blood-brain barrier choline transporter. Bioorg Med Chem Lett 20, 870–877.
Guangli, M., Yiyu, C. 2006. Predicting Caco-2 permeability using Support Vector Machine and Chemistry Development Kit. J Pharm Pharmaceut Sci 9, 210–221.
Gumbleton, M., Audus, K.L. 2001. Progress and limitations in the use of in vitro cell cultures to serve as permeability screen for the blood-brain barrier. J Pharm Sci 90, 1681–1698.
Hemmateenejad, B., Miri R., Safarpour, M.A., Mehdipour, A.R. 2006. Accurate prediction of the blood-brain partitioning of a large set of solutes using ab-initio calculations and genetic neural network modeling. J Comput Chem 27, 1125–1135.
Hou, T.J., Xu, X.J. 2003. ADME evaluation in drug discovery. 3. Modeling blood-brain barrier partitioning using simple molecular descriptors. J Chem Inf Comput Sci 43, 2137–2152.
Iyer, M., Mishru, R., Han, Y., Hopfinger, A.J. 2002. Predicting blood-brain barrier partitioning of organic molecules using membrane-interaction QSAR analysis. Pharm Res 19, 1611–1621.
Jolliet-Riant, P., Tillement, J.P. 1999. Drug transfer across the blood-brain barrier and improvement of brain delivery. Fundam Clin Pharmacol 13, 16–26.
Katritzky, A.R., Kuanar, M., Slavov, S., Dobchev, D.A., Fara, D.C., Karelson, M., Acree Jr., W.E., Solov’ev, V.P., Varnek, A. 2006. Correlation of bloodbrain penetration using structural descriptors. Bioorg. Med Chem 14, 4888–4917.
Kelder, J., Grootenhuis, P.D., Bayada D.M., Delbressine, L.P., Ploemen, J.P. 1999. Polar molecular surface as a dominating determinant for oral absorption and brain penetration of drugs. Pharm Res 16, 1514–1519.
Keseru, G.M., Molnar, L. 2001. High-throughput prediction of blood-brain partitioning: A thermodynamic approach. J Chem Inf Comput Sci 41, 120–128.
Konovalov, D.A., Sim, N., Deconinck, E., Vander Heyden, Y., Coomans, D. 2008. Statistical confidence for variable selection via Monte Carlo cross-validation. J Chem Inf Model 48, 370–383.
Kortagere, S., Chekmarev, D., Welsh, J.W., Ekins, S. 2008. New predictive models for blood-brain barrier permeability of drug-like molecules. Pharm Res 25, 1836–1845.
Lanevskij, K., Japertas, P., Didziapetris, R., Petrauskas, A. 2009. Ionization-specific prediction of blood-brain permeability. J Pharm Sci 98, 122–134.
Lombardo, F., Blake, J.F., Curatolo, W.J. 1996. Computation of brain-blood partitioning of organic solutes via free energy calculations. J Med Chem 39, 4750–4755.
Luco, J.M. 1999. Prediction of the brain-blood distribution of a large set of drugs from structurally derived descriptors using partial least-squares (PLS) modeling. J Chem Inf Comput Sci 39, 396–404.
Ma, X.L., Chen, C., Yang, J. 2005. Predictive model of blood-brain barrier penetration of organic compounds. Acta Pharmacol Sin 26, 500–512.
Norinder, U., Haeberlein, M. 2002. Computational approaches to the prediction of the blood-brain distribution. Adv Drug Deliv Rev 54, 291–313.
Norinder, U., Sjoberg, P., Osterberg, T. 1998. Theoretical calculation and prediction of brain-blood partitioning of organic solutes using MolSurf parametrization and PLS statistics. J Pharm Sci 87, 952–959.
Pan, D., Iyer, M., Liu, J., Li, Y., Hopfinger, A.J. 2004. Constructing optimum blood brain barrier QSAR models using a combination of 4D-molecular similarity measures and cluster analysis. J Chem Inf Comput Sci 44, 2083–2098.
Pardridge, W.M. 2001. Drug targeting, drug discovery and brain drug development. In: Brain Drug Targeting: The Future of Brain Drug Development, Cambridge University Press, Los Angeles, 1–12.
Pardridge, W.M. 2003. Blood-brain barrier drug targeting: the future of brain drug development. Mol Interv 3, 90–105.
Platts, J.A., Abraham, M.H., Zhao, Y.H., Hersey, A., Ijaz, L., Butina, D. 2001. Correlation and prediction of a large blood-brain distribution data set-an LFER study. Eur J Med Chem 36, 719–730.
Rose, K., Hall, L.H., Kier, L.B. 2002. Modeling bloodbrain barrier partitioning using the electrotopological state. J Chem Inf Comput Sci 42, 651–666.
Salminen, T., Pulli, A., Taskinen, J. 1997. Relationship between immobilised artificial membrane chromatographic retention and the brain penetration of structurally diverse drugs. J Pharm Biomed Anal 15, 469–477.
Scala, S., Akhmed, N., Rao, U.S., Paull, K., Lan, L.B., Dickstein, B., Lee, J.S., Elgemeie, G.H., Stein, W.D., Bates, S.E. 1997. P-glycoprotein substrates and antagonists cluster into two distinct groups. Mol Pharmacol 51, 1024–1033.
Smith, Q.R. 2003. A review of blood-brain barrier transport techniques. Methods Mol Med 89, 193–208.
Subramanian, G., Kitchen, D.B. 2003. Computational models to predict blood-brain barrier permeation and CNS activity. J Comput Aided Mol Des 17, 643–664.
van de Waterbeemd, H., Gifford, E. 2003. ADMET in silico modelling: Towards prediction paradise. Nat Rev Drug Discov 2, 192–204.
Vilar, S., Chakrabarti, M., Costanzi, S. 2010. Prediction of passive blood-brain partitioning: Straightforward and effective classification models based on in silico derived physicochemical descriptors. J Mol Graph Model 28, 899–903.
Winkler, D.A., Burden, F.R. 2004. Modelling bloodbrain barrier partitioning using Bayesian neural networks. J Mol Graph Model 22, 499–505.
Yang, S.Y., Huang, Q., Li, L.L., Ma, C.Y., Zhang, H., Bai, R., Teng, Q.Z., Xiang, M.L., Wei, Y.Q. 2009. An integrated scheme for feature selection and parameter setting in the support vector machine modeling and its application to the prediction of pharmacokinetic properties of drugs. Artif Intel Med 46, 155–163.
Young, R.C., Mitchell, R.C., Brown, T.H., Ganellin, C.R., Griffiths, R., Jones, M., Rana, K.K., Saunders, D., Smith, I.R., Sore, N.E., Wilks, T.J. 1988. Development of a new physico-chemical model for brain penetration and its application to the design of centrally acting H2 receptor histamine antagonists. J Med Chem 31, 656–671.
Zhang, L., Zhu, H., Oprea, I.T., Golbraikh, A., Tropsha, A. 2008. QSAR modeling of the blood-brain barrier permeability for diverse organic compounds. Pharm Res 25, 1902–1914.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kumar, R., Sharma, A. & Tiwari, R.K. Can we predict blood brain barrier permeability of ligands using computational approaches?. Interdiscip Sci Comput Life Sci 5, 95–101 (2013). https://doi.org/10.1007/s12539-013-0158-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-013-0158-9