Abstract
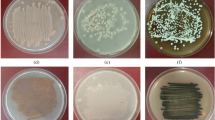
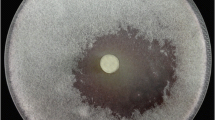
Thirty soil-isolates of Streptomyces were analyzed to determine their antagonism against plant-pathogenic fungi including Fusarium oxysporum, Pythium aristosporum, Colletotrichum gossypii, and Rhizoctonia solani. Seven isolates showed antifungal activity against one or more strain of the tested fungi. Based on the 16S rDNA sequence analysis, these isolates were identified as Streptomyces tendae (YH3), S. griseus (YH8), S. variabilis (YH21), S. endus (YH24), S. violaceusniger (YH27A), S. endus (YH27B), and S. griseus (YH27C). The identity percentages ranged from 98 to 100%. Although some isolates belonged to the same species, there were many differences in their cultural and morphological characteristics. Six isolates out of seven showed chitinase activity according to a chitinolytic activity test and on colloidal chitin agar plates. Based on the conserved regions among the family 19 chitinase genes of Streptomyces sp. two primers were used for detection of the chitinase (chiC) gene in the six isolates. A DNA fragment of 1.4 kb was observed only for the isolates YH8, YH27A, and YH27C. In conclusion, six Streptomyces strains with potential chitinolytic activity were identified from the local environment in Taif City, Saudi Arabia. Of these isolates, three belong to family 19 chitinases. To our knowledge, this is the first reported presence of a chiC gene in S. violaceusniger YH27A.
Similar content being viewed by others
References
Al-Kahtani, M., AI-Khalil, A., AI-Kadeeb, S., and Hassan, H.Z. 2008. Molecular genetic fingerprint of some Streptomyces isolated from Riyadh city. Saudi J. Biological Sci. 15, 243–251.
Blaak, H., Schnellmann, J., Walter, S., Henrissat, B., and Schrempf, H. 1993. Characteristics of an exochitinase from Streptomyces olivaceoviridis, its corresponding gene, putative protein domains and relationship to other chitinases. Eur. J. Biochem. 214, 659–669.
Bonjar, S., Fooladi, M.H., Mahdavi, M.J., and Shahghasi, A. 2004. Broadspectrim, a novel antibacterial from Streptomyces sp. Biotechnol. 3, 126–130.
Bradford, M.M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254.
Collinge, D.B., Kragh, K.M., Mikkelsen, J.D., Nielsen, K.K., Rasmussen, U., and Vad, K. 1993. Plant chitinases. Plant J. 3, 31–40.
Davies, G. and Henrissat, B. 1995. Structures and mechanisms of glycosyl hydrolases. Structure 3, 853–859.
Errakhi, R., Bouteau, F., Lebrihi, A., and Barakate, M. 2010. Evidences of biological control capacities of Streptomyces spp. against Sclerotium rolfsii responsible for damping-off disease in sugar beet (Beta vulgaris L.). World J. Microbiol. Biotechnol. 23, 1503–1509.
Fujii, T. and Miyashita, K. 1993. Multiple domain structure in a chitinase gene (chiC) of Streptomyces lividans. J. Gen. Microbiol. 139, 677–686.
Heitz, T., Segond, S., Kaumann, S., Georoy, P., Prasad, V., Brunner, F., Fritig, B., and Legrand, M. 1994. Molecular characterization of a novel tobacco pathogenesis-related (PR) protein: a new plant chitinase/lysozyme. Mol. Gen. Genet. 245, 246–254.
Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280, 309–316.
Henrissat, B. and Bairoch, A. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293, 781–788.
Hoster, F., Schmitz, J.E., and Daniel, R. 2005. Enrichment of chitinolytic microorganisms: isolation and characterization of a chitinase exhibiting antifungal activity against phytopathogenic fungi from a novel Streptomyces strain. Appl. Microbiol. Biotechnol. 66, 434–442.
Itoh, Y., Kawase, T., Nikaidou, N., Fukada, H., Mitsutomi, M., Watanabe, T., and Itoh, Y. 2002. Functional analysis of the chitin-binding domain of a family 19 chitinase from Streptomyces griseus HUT6037: substrate-binding affinity and cis-dominant increase of antifungal function. Biosci. Biotechnol. Biochem. 66, 1084–1092.
Kawase, T., Saito, A., Sato, T., Kanai, R., Fujii, T., Nikaidou, N., Miyashita, K., and Watanabe, T. 2004. Distribution and phylogenetic analysis of family 19 chitinases in Actinobacteria. Appl. Environ. Microbiol. 70, 1135–1144.
Kawase, T., Yokokawa, S., Saito, A., Fujii, T., Nikaidou, N., Miyashita, K., and Watanabe, T. 2006. Comparison of enzymatic and antifungal properties between family 18 and 19 chitinases from S. coelicolor A3 (2). Biosci. Biotechnol. Biochem. 70, 988–998.
Keiser, T., Bibb, M.J., Buttner, M.J., Chater, K.F., and Hopwood, D.A. 2000. General introduction to actinomycete biology, pp. 1–21. In Practical Streptomyces genetics. The John Innes Foundation, England.
Kim, K. and Ji, H.-S. 2001. Effect of chitin sources on production of chitinase and chitosanase by Streptomyces griseus HUT 6037. Biotechnol. Bioprocess Eng. 6, 18–24.
Kimura, M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120.
Leah, R., Tommerup, H., Svendsen, I., and Mundy, J. 1991. Biochemical and molecular characterization of three barley seed proteins with antifungal properties. J. Biol. Chem. 266, 1564–1573.
McSpadden-Gardener, B.B. and Fravel, D.R. 2002. Biological control of plant pathogens: Research, commercialization and application in the USA. http://www.apsnet.org/online/feature/biocontrol/to p.html.
Miyashita, K. and Fujii, T. 1993. Nucleotide sequence and analysis of a gene (chiA) for chitinase from Streptomyces lividans 66. Biosci. Biotechnol. Biochem. 57, 1691–1698.
Miyashita, K., Fujii, T., Watanabe, W., and Ueno, H. 1997. Nucleotide sequence and expression of a gene (chiB) for a chitinase from Streptomyces lividans. J. Ferment. Bioeng. 83, 26–31.
Miyashita, K., Fujii, T., and Sawada, Y. 1991. Molecular cloning and characterization of chitinase genes from Streptomyces lividans 66. J. Gen. Microbiol. 137, 2065–2072.
Narayana, K.J.P. and Vijayalakshmi, M. 2009. Chitinase production by Streptomyces sp. Anu 6277. Braz. J. Microbiol. 40, 725–733.
Ohno, T., Armand, S., Hata, T., Nikaidou, N., Henrissat, B., Mitsutomi, M., and Watanabe, T. 1996. A modular family 19 chitinase found in the prokaryotic organism Streptomyces griseus HUT 6037. J. Bacteriol. 178, 5065–5070.
Omura, S. 1986. Philosophy of new drug discovery. Microbiol. Rev. 50, 259–279.
Ordentlich, A., Elad, Y., and Chet, I. 1988. The role of chitinase of Serratia marcescens in biocontrol of Sclerotium rolfsii. Phytopathology 787, 84–88.
Park, H.S., John, J., and Kilbane, I.I. 2006. Rapid detection and high-resolution discrimination of the genus Streptomyces based on 16S rRNA spacer region and denaturing gradient gel electrophoresis. J. Ind. Microbiol. Biotechnol. 33, 289–297.
Patil, R.S., Ghormade, V., and Deshpande. M.V. 2000. Chitinolytic enzymes: an expoloration. Enzyme Microb. Technol. 26, 473–483.
Robbins, P.W., Albright, C., and Benfield, B. 1988. Cloning and expression of a Streptomyces plicatus chitinase (chitinase-63) in Escherichia coli. J. Biol. Chem. 263, 443–447.
Robbins, P.W., Oberbye, K., Albright, C., Benfield, B., and Pero, J. 1992. Cloning and high-level expression of chitinase-encoding gene of Streptomyces plicatus. Gene 111, 69–76.
Roberts, W.K. and Selitrennikoff, C.P. 1986. Isolation and partial characterization of two antifungal proteins from barley. Biochim. Biophys. Acta 880, 161–170.
Saito, A., Fujii, T., Yoneyama, T., and Miyashita, K. 1998. glkA is involved in glucose repression of chitinase production in Streptomyces lividans. J. Bacteriol. 180, 2911–2914.
Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., USA.
Shanmugaiah, V., Mathivanan, N., Balasubramanian, N., and Manoharan, P.T. 2008. Optimization of cultural conditions for production of chitinase by Bacillus laterosporous MML2270 isolated from rice rhizosphere soil. Afr. J. Biotechnol. 7, 2562–2568.
Shirling, E.B. and Gottlieb, D. 1966. Methods for characterization of Streptomyces species. Inst. J. Syst. Bacteriol. 16, 313–340.
Suresh, P.V. and Chandrasekaran, M. 1998. Utilization of prawn waste for chitinase production by the marine fungus Beauveria bassiana by solid state fermentation. World J. Microbiol. Biotechnol. 14, 655–660.
Taechowisan, T., Peberdy, F.P., and Lumyong, S. 2004. PCR cloning and heterologous expression of chitinase gene of endophytic Streptomyces aureofaciens CMUAc130. J. Gen. Appl. Microbiol. 50, 177–182.
Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24, 4876–4882.
Tripathi, G. and Rawal, S.K. 1998. Simple and efficient protocol for isolation of high molecular weight DNA from Streptomyces aureofaciens. Biotechnol. Tech. 12, 629–631.
Tsujibo, H., Endo, H., Minoura, K., Miyamoto, K., and Inamori, Y. 1993. Cloning and sequence analysis of the gene encoding a thermostable chitinase from Streptomyces thermoviolaceus OPC-520. Gene 134, 113–117.
Tsujibo, H., Okamoto, T., Hatano, N., Miyamoto, K., Watanabe, T., Mitsutomi, M., and Inamori, Y. 2000. Family 19 chitinases from Streptomyces thermoviolaceus OPC-520: Molecular cloning and characterization. Biosci. Biotechnol. Biochem. 64, 2445–2453.
Vyas, P. and Deshpande, M.V. 1989. Chitinase production by Myrothecium verrucaria and its significant for fungal mycelia degradation. J. Gen. Appl. Microbiol. 35, 343–350.
Vyas, P. and Deshpande, M.V. 1991. Enzymatic hydrolysis of chitin by Myrothecium verrucaria chitinase complex and its utilization to produce SCP. J. Gen. Appl. Microbiol. 37, 267–275.
Watanabe, T., Kanai, R., Kawase, T., Tanabe, T., Misutomi, M., Sakuda, S., and Miyashita, K. 1999. Family 19 chitinase of Streptomyces species: characterization and distribution. Microbiology 145, 3353–3363.
Weisburg, W.G., Barns, S.M., Pelletier, D.A., and Lane, D.J. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703.
Weisburg, W.G., Tully, J.G., Rose, D.L., Petzel, J.P., Oyaizu, H., Yang, D., Mandelco, L., Sechrest, J., Lawrence, T.G., Van Etten, J., and et al. 1989. A phylogenetic analysis of the mycoplasmas: basis for their classification. J. Bacteriol. 171, 6455–6467.
Williams, S.T., Goodfellow, M., Alderson, G., Wellington, E.M.H., Sneath, P.H.A., and Sackin, M. 1983. Numerical classification of Streptomyces and related genera. J. Gen. Microbiol. 129, 1743–1813.
Xiao, K., Kinkel, L.L., and Samac, D.A. 2002. Biological control of phytophthora root rots on alfalfa and soybean with Streptomyces. Biol. Control. 23, 285–295.
Yücel, S. and Yamaç M. 2010. Selection of Streptomyces isolates from turkish karstic caves against antibiotic resistant microorganisms. Pak. J. Pharm. Sci. 23, 1–6.
Zarandi, M.E., Shahidi Bonjar, G.H., Dehkaei, F.P., Moosavi, S.A.A., Farokhi, P.R., and Aghighi, S. 2009. Biological control of rice blast (Magnaporthe oryzae) by use of Streptomyces sindeneusis isolate 263 in greenhouse. Am. J. Appl. Sci. 6, 194–199.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gherbawy, Y., Elhariry, H., Altalhi, A. et al. Molecular screening of Streptomyces isolates for antifungal activity and family 19 chitinase enzymes. J Microbiol. 50, 459–468 (2012). https://doi.org/10.1007/s12275-012-2095-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-012-2095-4