Abstract
We analysed single nucleotide polymorphisms (SNPs) tagging the genetic variability of six candidate genes (ATF6, FABP1, LPIN2, LPIN3, MLXIPL and MTTP) involved in the regulation of hepatic lipid metabolism, an important regulatory site of energy balance for associations with body mass index (BMI) and changes in weight and waist circumference. We also investigated effect modification by sex and dietary intake. Data of 6,287 individuals participating in the European prospective investigation into cancer and nutrition were included in the analyses. Data on weight and waist circumference were followed up for 6.9 ± 2.5 years. Association of 69 tagSNPs with baseline BMI and annual changes in weight as well as waist circumference were investigated using linear regression analysis. Interactions with sex, GI and intake of carbohydrates, fat as well as saturated, monounsaturated and polyunsaturated fatty acids were examined by including multiplicative SNP-covariate terms into the regression model. Neither baseline BMI nor annual weight or waist circumference changes were significantly associated with variation in the selected genes in the entire study population after correction for multiple testing. One SNP (rs1164) in LPIN2 appeared to be significantly interacting with sex (p = 0.0003) and was associated with greater annual weight gain in men (56.8 ± 23.7 g/year per allele, p = 0.02) than in women (−25.5 ± 19.8 g/year per allele, p = 0.2). With respect to gene–nutrient interaction, we could not detect any significant interactions when accounting for multiple testing. Therefore, out of our six candidate genes, LPIN2 may be considered as a candidate for further studies.
Similar content being viewed by others
Introduction
During the last decades, the prevalence of obesity has risen (Low et al. 2009) and a strong rise in prevalence of obesity with increasing age is observed (von Ruesten et al. 2011). Factors that influence weight gain during adulthood are incompletely understood, but it is believed that a person’s susceptibility to gain weight is influenced by an interplay of many factors including lifestyle factors like physical activity and nutritional habits as well as genetic factors (WHO 2000).
From the physiological view, weight gain results from a positive energy balance, where excessive energy is stored in form of fat in the adipose tissue. The liver is the central site of fatty acid and lipid metabolism and plays a regulatory role in energy homoeostasis. Impaired regulation of hepatic lipid metabolism may result in obesity (Guillou et al. 2008).
The activating transcription factor 6 (ATF6), liver fatty acid binding protein 1 (FABP1), lipin-2 (LPIN2), lipin-3 (LPIN3), carbohydrate response element binding protein (MLXIPL) and microsomal triglyceride transfer protein (MTTP) genes were considered to be candidate genes, and their function in relation to hepatic lipid metabolism is described as follows.
-
ATF6 is an endoplasmic reticulum (ER) membrane-bound transcription factor that is activated by ER stress. ER stress can be caused among other stress factors by metabolic stress, like glucose deprivation (Salminen and Kaarniranta 2010; Zeng et al. 2004). Upon activation, ATF6 regulates target genes for the unfolded protein response (Jäger et al. 2012) as well as several lipogenic genes (Zeng et al. 2004). It has been shown to antagonise the activity of sterol responsive element binding protein on cholesterol biosynthesis (Zeng et al. 2004).
-
FABP1 serves as an intracellular acceptor of long-chain fatty acids (McArthur et al. 1999) and is involved in the partitioning of fatty acids to specific lipid metabolic pathways (Veerkamp and van Moerkerk 1993; Atshaves et al. 2010). The FABP1 knockout mouse was shown to be protected against diet-induced obesity (Newberry et al. 2006).
-
The lipin proteins act as phosphatidate phosphatase-1 converting phosphatidate to diacylglycerol in triglyceride and phospholipid biosynthesis (Gropler et al. 2009). Lipin 2 mRNA and protein expression are induced by food deprivation (Gropler et al. 2009). Furthermore, lipin-2 and potentially lipin-3 function as transcriptional coactivators for peroxisome proliferator-activated receptor-response elements thereby promoting fatty acid oxidation (Donkor et al. 2009).
-
The carbohydrate response element binding protein (MLXIPL) regulates transcriptionally key enzymes of glycolysis and lipogenesis in the liver in a glucose-dependent manner (Iizuka and Horikawa 2008; Poupeau and Postic 2011).
-
MTTP is involved in the assembly and secretion of VLDLs in the liver (Gordon and Jamil 2000), and its expression is regulated by changes in the dietary intake of sugar and fat (Hussain et al. 2011).
Previous genetic studies have revealed associations of SNPs in these candidate genes anthropometric measures, i.e. body mass index (BMI) and fat mass (ATF6 (Fougeray et al. 2011), FABP1 (Weickert et al. 2007; Brouillette et al. 2004), LPIN2 (Aulchenko et al. 2007), MTTP (Ledmyr et al. 2002; Bohme et al. 2008; Berthier et al. 2004; Rubin et al. 2006)) as well as with lipid levels [ATF6 (Meex et al. 2009), FABP1 (Fisher et al. 2007; Brouillette et al. 2004), MLXIPL (Talmud et al. 2009; Kooner et al. 2008; Willer et al. 2008), MTTP (Bohme et al. 2008; Berthier et al. 2004; Ledmyr et al. 2002)] and type two diabetes [ATF6 (Meex et al. 2007; Thameem et al. 2006), FABP1 (Mansego et al. 2012), LPIN2 (Aulchenko et al. 2007; Parker et al. 2001)), MTTP (Rubin et al. 2006)].
Here, we aimed to study the association of variation in the selected candidate genes with changes in body weight and waist circumference. Since some previously reported associations of our candidate genes were sex specific (Bohme et al. 2008; Fisher et al. 2007), we further tested for interactions with sex. In addition, a major aim was to investigate possible interaction with dietary factors. As dietary effect modifiers, a quantitative (carbohydrate intake) and a qualitative measure [glycaemic index (GI)] of carbohydrate intake were chosen, since lipogenesis and lipolysis are mainly regulated by the intake of carbohydrates (Flatt 1995). Furthermore, interaction analyses were extended to dietary fat intake, since some of the genes have been shown to be sensitive to dietary fat with regard to regulation (Hussain et al. 2011) and interactions (Robitaille et al. 2004), respectively. Interaction and association analyses of the six selected candidate genes and baseline BMI, changes in body weight and waist circumference were carried out in a large prospective cohort from five European populations.
Methods
Study population
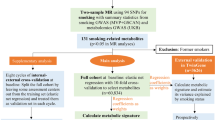
The study is based on several existing cohorts of the European prospective investigation into cancer and nutrition (EPIC) (Riboli and Kaaks 1997; Riboli et al. 2002) that participated in the DiOGenes project (www.diogenes-eu.org; Saris and Harper 2005) with the aim to study dietary components that influence weight gain in combination with genetic and behavioural factors. The EPIC study has been described in detail earlier (Riboli and Kaaks 1997; Riboli et al. 2002). Data from the cohorts in eight regions in five different European countries were included into the DiOGenes study: Copenhagen and Aarhus (Denmark), Potsdam (Germany), Florence (Italy), Bilthoven (combined EPIC centre from subcentres located in Doetinchem, Amsterdam and Maastricht; The Netherlands) and Norfolk (United Kingdom). Together, these cohorts comprise 146,543 individuals. Participants were considered eligible for the current analyses if they were younger than 60 years at baseline and younger than 65 years at follow-up, had blood samples available and had baseline information on diet, weight and height and follow-up information on weight. Inclusion criteria were also stable smoking habits, no diagnosis of cancer, cardiovascular disease and diabetes at baseline or during follow-up, and an annual weight change not higher than 5 kg/year. A total of 50,293 individuals were eligible. A subcohort was drawn randomly from the total eligible cohort as described previously (Du et al. 2011; Vimaleswaran et al. 2012; Fisher et al. 2012). The final subcohort including 7,061 individuals was used for the current analysis.
Anthropometrics, dietary intake and other measurements
Baseline weight, height and waist circumference were assessed by trained staff according to a predefined protocol (Rinaldi et al. 2006). During follow-up, participants in Doetinchem (Netherlands) and Norfolk (UK) were re-examined using the same protocol while in all other centres self-reported data was collected.
Validated country-specific food frequency questionnaires were completed by participants during the baseline assessment (Riboli and Kaaks 1997). Energy and nutrient intakes were calculated using country-specific food composition tables (Riboli and Kaaks 1997). Dietary GI was calculated using glucose as the reference food according to a methodology described elsewhere (van Bakel et al. 2009; Du et al. 2008). Data on lifestyle were collected using self-administered questionnaires at baseline and follow-up.
Selection and genotyping of candidate genes and SNPs
We have selected candidate genes based on their involvement in or regulation of lipid metabolism, expression in the liver, a potential nutrient-sensitive effect and previous reported genetic association with weight or body composition in human (Fougeray et al. 2011; Weickert et al. 2007; Aulchenko et al. 2007; Berthier et al. 2004; Bohme et al. 2008; Brouillette et al. 2004; Ledmyr et al. 2002; Rubin et al. 2006) or animal studies (He et al. 2009).
Tagging SNPs in the selected candidate genes were chosen based on data from the International HapMap project (The International HapMap Consortium 2005) using pairwise tagging and R 2–threshold of 0.8 within the Tagger software (de Bakker et al. 2005) implemented in Haploview V3.3 (Barrett et al. 2005) to cover common genetic variation (MAF >0.05). SNPs were forced to be included when they had potential or proven functional implications or had been extensively studied in the past. High throughput genotyping of a total of 86 SNPs in the ATF6, FABP1, LPIN2, LPIN3, MLXIPL and MTTP genes was performed using an Illumina Beadstation Genotyping System along with other SNPs that served different genetic analyses within the DiOGenes project. The initial stage of quality control stratified by study centre was carried out jointly for all SNPs. Genotyping (i.e. individuals) passed the initial stage of quality control if their discordance rate of duplicate samples was ≤3 % and at least 95 % of SNPs per sample were genotyped. SNPs were excluded unless ≥95 % of samples were genotyped. Furthermore, SNPs were excluded for a particular country when the corresponding p value for Hardy–Weinberg equilibrium was below 0.001. Genotype data of 6,566 (93.0 %) individuals passed quality control. From the 86 SNPs that were selected for this study, seven SNPs did not pass the initial stage of quality control (Online Resource 1-Table S1). Another ten SNPs had to be further excluded from the analyses because they only passed quality control in some, but not all, EPIC-centres (Online Resource 1-Table S1). Eventually, a total of 69 SNPs were included in the current analyses.
Linkage disequilibrium (LD)
LD between genotyped variants within the study population was calculated using Haploview V3.3 (Barrett et al. 2005). LD of genotyped LPIN2 variants and rs3745012 variant reported in literature (Aulchenko et al. 2007) was established using SNAP and the 1000GenomesPilot1 dataset (Johnson et al. 2008; Abecasis et al. 2010).
Statistical analyses
All analyses were carried out separately for each country, and combined effect estimates with corresponding p values were calculated using random effects meta-analysis. Due to differences in data collection of anthropometric measures and length of follow-up, data of the Dutch cohort was split into two study centres resulting in the final allocation of six study centres, namely Copenhagen and Århus (Denmark, DK-CopAa), Potsdam (Germany, GER-Pot), Florence (Italy, IT-Flo), Doetinchem (Netherlands, NL-Doe), Amsterdam and Maastricht (Netherlands, NL-AmMa), and Norfolk/England (United Kingdom, UK-Nor).
Linear regression models were applied assuming an additive genetic model. Analyses of baseline BMI were adjusted for baseline values of age, sex, height, smoking status, energy intake, physical activity and recruitment centre. Annual weight change was calculated by subtracting baseline body weight from follow-up body weight and dividing the difference in grams by the duration of follow-up in years (y). Analyses of annual weight change were adjusted for baseline values of age, sex, height, smoking status, weight, energy intake, physical activity and recruitment centre and follow-up time. Analyses of annual changes in waist circumference were adjusted for baseline age, sex, height, smoking status, waist circumference, energy intake, physical activity and recruitment centre and follow-up time. Interaction with sex was assessed by including the multiplicative interaction term of sex (indicator variable; 0/1) and each SNP in the model. When the interaction terms were statistically significant, sex-stratified analyses were carried out.
Interaction with carbohydrate intake was assessed by a multiplicative interaction term of the percentage of energy derived from carbohydrates and each SNP in a linear model that was adjusted for percentages of energy derived from carbohydrates, protein and alcohol (all scaled by units corresponding to 5 % of total energy intake), the ratio of monounsaturated to saturated and the ratio of polyunsaturated to saturated fatty acids in addition to the basic set of covariates described above. Interactions with total fat intake were modelled in the same way with additional adjustment of energy derived from fat, protein, alcohol as well as the fatty acid ratios in addition to the basic set of covariates. Fat-subtype-based interaction analyses of saturated, monounsaturated and polyunsaturated fat, respectively, were modelled like total fat but additionally adjusted for the complementary fat intake, i.e., e.g. when saturated fat is used, it is also adjusted for the difference between total fat intake and saturated fat intake. Interactions between SNPs and GI were tested by adding the multiplicative interaction term of GI and each SNP as well as the GI main effect to the basic linear regression model for waist and weight gain. All statistical analyses were carried using Stata 9.2 for Windows (StataCorp LP, Texas, USA; www.stata.com). The forest plot was drawn using R version 2.15.1 (R Development Core Team 2010) and the ‘meta’ package (Schwarzer 2010).
The Bonferroni method was used with respect to each analysis case to correct for the multiple testing of 69 SNPs. Therefore, a p value lower than 0.0007 (0.05/69) was considered to be statistically significant. Power calculations were performed using the QUANTO software Version 1.2.4 (May 2009; Gauderman and Morrison 2006). The minimum detectable SNP effects with respect to different minor allele frequencies (MAFs) assuming a 80 % power rate and for a given significance level of α = 0.05 were estimated to range from 40 g/year (MAF = 50 %) to 88 g/year (MAF = 5 %) for weight gain, from 0.18 kg/m2 (MAF = 50 %) to 0.40 kg/m2 (MAF = 5 %) for BMI and from 0.06 cm/y (MAF = 50 %) to 0.14 cm/y (MAF = 5 %) for waist circumference.
Results
Study population characteristics
The final study population included 6,287 individuals and consisted of 46 % men. The mean ± SD baseline age and BMI were 48.0 ± 7.2 year and 25.4 ± 3.7 kg/m2, respectively. Individuals had a mean follow-up time of 6.9 ± 2.5 year, where they gained on average 235 ± 784 g body weight per year (g/year) and 0.7 ± 1.2 cm waist circumference per year (cm/year), respectively. Age and follow-up time were similar between men and women (Table 1). Annual changes in weight and waist circumference were slightly higher in women than in men (Table 1). A list of successfully genotyped SNPs and their MAFs can be found in Online Resource 1-Table S2.
In all genes, a number of nominally significant p values were observed, mainly in the interaction analyses and to a lesser extent in the main effects analyses (Table 2; detailed results in Online Resource 1-Table S3-11). Nominally significant main effects of FABP1 rs894194 and LPIN2 rs10438875 on baseline BMI as well as of MLXIPL rs6967028 and two SNPs in MTTP (rs1057613, rs3816873) on annual weight change were detected (Table 2).
Only the interaction of the LPIN2 SNP rs1164 with sex in association with weight change was significant after adjustment for multiple testing (β = −111.5 ± 30.9 g/year, p = 0.0003). For this SNP, sex-stratified analyses were carried out. As shown in Fig. 1, the presence of the minor allele of LPIN2 rs1164 is estimated to lead to a weight gain of 56.8 ± 23.7 g/year per minor allele in men (p = 0.02) whereas in women, a small weight loss was observed (β = −25.5 ± 19.8 g/year per minor allele, p = 0.2). The study-specific effect estimates in both men and women varied between studies (Fig. 1), but were consistently larger in men than in women throughout.
All tested gene–diet interactions were statistically not significant after correction for multiple testing. The lowest p values for gene–diet interactions were observed for firstly, an interaction of rs2919871 in FABP1 and intake of saturated fat in association with weight change (β = 81.4 ± 24.9 g/year, p = 0.001) and secondly, an interaction of SNP rs3934427 in LPIN2 with intake of carbohydrates in association with weight change (β = 44.1 ± 14.3 g/year, p = 0.002). Both SNPs showed further nominally significant gene–diet interactions; the FABP1 rs2919871 variant with total fat (p = 0.04) again in association with weight gain. The LPIN2 rs3934427 SNP presented the same interaction with dietary carbohydrates in association with waist change (p = 0.01) as well as an interaction with the intake of fat in association with waist change (p = 0.05).
Discussion
For 69 SNPs located in the six candidate genes (ATF6, FABP1, LPIN2, LPIN3, MLXIPL and MTTP), no significant associations with baseline BMI, annual change in waist circumference or body weight as well as no interactions with dietary factors (carbohydrate intake, GI, total, saturated, monounsaturated and polyunsaturated fat) were detected following correction for multiple testing. Altogether, only the interaction of rs1164 in LPIN2 with sex on annual weight change can be considered statistically significant taking multiple testing into account.
Strength and limitations
Our study investigated the interaction of common genetic variation in proteins involved in lipid uptake and metabolism of the liver and dietary factors (i.e. carbohydrates, fat and GI) in relation to weight development in five European populations. Large prospective cohort studies like this which offer detailed dietary records of their participants are still rare. Therefore, this study seems to be unique to investigate gene–nutrient interactions, but there are also important limitations. First, the study includes the possibility of some degree of under- and misreporting of weight and waist changes as well as of dietary intake. For all, but two centres, body weight and waist circumference were measured in the study centre by trained staff at baseline, while the follow-up measurement was self-reported. In the remaining two centres (UK-Nor, NL-Doe), body weight and waist circumference were also taken in the study centre during follow-up. Anthropometric measurements are usually underreported by self-reports (Park et al. 2011). Therefore, our data on weight gain based on measured baseline values and self-reported follow-up values would underestimate the true weight gain and could therefore attenuate the observed effect. Furthermore, the degree of underreporting might vary according to gender (Park et al. 2011), which could create an artificial interaction with sex. In the centre-specific analysis, the overall significant LPIN2 rs1164–sex interaction was nominally significant only in two centres with self-reports (IT-Flo and NL-AmMa), but in none with measured follow-up data (UK-Nor, NL-Doe). However, the estimate for the interaction effect in the British centre with measured follow-up data (UK-Nor −136.8 ± 107.1 g/year, p = 0.20) is of the same magnitude as in the two centres with self-reported follow-up data and significant interaction effect (It-Flo: −124.5 ± 59.9 g/year, p = 0.04; NL-AmMa: −175.7 ± 82.6 g/year, p = 0.03) suggesting that differences in data collection had no major influence on the observed effect. Second, while the genotype is a stable exposure, dietary intake may change over time, especially as a result of weight changes. We have analysed dietary intake only once at baseline and are therefore not able to take changes in dietary intake into account. Third, the selection of candidate genes was based on prior knowledge and financial resources and represents a small subset of the ‘key players’ within hepatic fat metabolism. We therefore have to acknowledge that, since the regulation of hepatic lipid metabolism is much more complex, other relevant genetic variants among these pathways might have been missed. Fourth, although our study is large, we cannot exclude the possibility that the lack of significant findings is due to a lack of power. Singular genetic effects in polygenic traits like weight gain are supposed to be small, and the power to detect an interaction with dietary intake is further limited by the measurement error in estimating dietary intake. Moreover, the Bonferroni method was used to correct for multiple testing. The Bonferroni method is frequently criticised for being relative strict and producing false negative results (Perneger 1998). However, since in most cases p values were just below the nominally significance threshold of 0.05, less conservative methods would most likely not have changed the overall result. However, since our study is based on meta-analyses of data from five different countries, a common type I error (rejection of null hypothesis that is actually true) seemed not very likely to have occurred. For that reason, nominally significant results are discussed in the following section provided they have been reported in the literature previously.
Findings
The significant SNP rs1164–sex interaction resulted from a larger annual weight gain in men than in women. In the past, the rs3745012 LPIN2 variant has been described to be associated with BMI, waist circumference and other anthropometric measurements in a Dutch study (Aulchenko et al. 2007). However, both variants are not in LD (R 2 = 0.05). All other variants in LD (R 2 = 0.77) with rs3745012 were not associated with any of the analysed traits. Therefore, we feel confident to exclude rs3745012 as a potential variant underlying the observed association.
The detected interaction effect led to weight gain in men, whereas in women, a small weight loss was observed. A recent GWAS meta-analysis of sex-specific genetic effects for height, weight, body mass index, waist circumference, hip circumference and waist-to-hip ratio did not find a sex-specific effect in LPIN2 (Randall et al. 2013). Therefore, our finding needs to be treated very cautiously.
Lipin 2 catalyses as phosphatidate phosphatase a key step of lipid biosynthesis and regulates fatty acid oxidation as transcriptional coactivator of PPARα as well as PPARγ coactivator 1a (Reue and Dwyer 2009). Rare mutations in LPIN2 cause the auto-inflammatory disorder Majeed syndrome suggesting a role in regulation of the inflammatory response for LPIN2 (Ferguson et al. 2005; Reue and Donkor 2007). It seems to be possible to influence body weight regulation via these mechanisms; however, as the causal variant is unknown, it is impossible to elucidate a biological mechanism for the observed association.
Two SNPs (rs1057613, rs3816873 (I128T), R 2 = 0.41) in MTTP showed a nominally significant association with annual changes in body weight. MTTP plays a central role in lipoprotein assembly (Hussain et al. 2003). The T128 allele of rs3816873 has been reported to lead to reduced function of MTTP (Ledmyr et al. 2006). The SNP is in complete LD with several promoter variants of the gene that have been shown to alter MTTP expression (Rubin et al. 2008; Ledmyr et al. 2004; Aminoff et al. 2010; Karpe et al. 1998). The rs3816873 polymorphism was reported to be associated with BMI and other anthropometric measures, but results were inconsistent (Rubin et al. 2006; Berthier et al. 2004; St-Pierre et al. 2002; Ledmyr et al. 2002). The so far largest study of MTTP variants on BMI in 7582 KORA participants found no association of BMI and waist circumference with the rs3816873 polymorphism, but an association with the H297Q (rs2306985) polymorphism in women (Bohme et al. 2008). In the present study, both variants were not associated with BMI, but the T128 allele was associated with increased weight gain (β = 38.2 ± 16.5 g/year per minor allele, p = 0.02). Since it has been shown that the effect of MTTP on BMI was sex specific (Rubin et al. 2006; Berthier et al. 2004), sex-stratified analyses were carried out. Although no statistically significant interaction between sex and rs3816873 was detected, a nominally significant association of rs3816873 with increased annual weight gain was detected in men only (men: β = 57.9 ± 19.9 g/year per minor allele, p = 0.004; women: β = 17.8 ± 17.6 g/year per minor allele, p = 0.31). The so far contrasting findings might be explained by different nutritional influences. Here, we tested for interactions with the intake of carbohydrates, GI, fat and fat subtypes but did not detect any influence of those dietary factors on the association of SNPs in MTTP and weight change.
There have been previous reports on an association with BMI and waist circumference for the FABP1 T94A variant (rs2241883) (Brouillette et al. 2004; Weickert et al. 2007). In the present study, we did not observe an association with BMI or changes in body weight or waist circumference of this variant.
The DiOGenes project also included an intervention study that investigated weight regain after an initial weight loss with dietary intervention differing in GI and protein intake (Larsen et al. 2012). This intervention study analysed the same genes as our study for main effects of weight, waist and fat mass regain after weight loss and interaction with protein content and GI of the dietary intervention (Larsen et al. 2012). As in our study, no significant main or SNP–diet interaction effects were identified when correction for multiple testing was applied.
In summary, we found a significant interaction of rs1164 in LPIN2 with sex in association with weight gain, leading to higher weight gain in of about 90 g/year in men than in women (men: 56.8 ± 23.7 g/year per minor allele; women: −25.5 ± 19.8 g/year per minor allele) than in women. We observed no significant association between SNPs located in ATF6, FABP1, LPIN3, MTTP and MLXIPL and annual changes in weight and waist circumference after correction for multiple testing. Furthermore, we found no convincing evidence for interactions of these candidate genes and either intake of carbohydrates, GI, total, saturated, monounsaturated and polyunsaturated fat. From our six selected candidate genes, LPIN2 is the one that could be followed up in further studies with a special focus on sex-specific effects.
References
Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA (2010) A map of human genome variation from population-scale sequencing. Nature 467(7319):1061–1073. doi:10.1038/nature09534
Aminoff A, Ledmyr H, Thulin P, Lundell K, Nunez L, Strandhagen E, Murphy C, Lidberg U, Westerbacka J, Franco-Cereceda A, Liska J, Nielsen LB, Gafvels M, Mannila MN, Hamsten A, Yki-Jarvinen H, Thelle D, Eriksson P, Boren J, Ehrenborg E (2010) Allele-specific regulation of MTTP expression influences the risk of ischemic heart disease. J Lipid Res 51(1):103–111. doi:10.1194/jlr.M900195-JLR200
Atshaves BP, Martin GG, Hostetler HA, McIntosh AL, Kier AB, Schroeder F (2010) Liver fatty acid-binding protein and obesity. J Nutr Biochem 21(11):1015–1032. doi:10.1016/j.jnutbio.2010.01.005
Aulchenko YS, Pullen J, Kloosterman WP, Yazdanpanah M, Hofman A, Vaessen N, Snijders PJLM, Zubakov D, Mackay I, Olavesen M, Sidhu B, Smith VE, Carey A, Berezikov E, Uitterlinden AG, Plasterk RHA, Oostra BA, van Duijn CM (2007) LPIN2 is associated with type 2 diabetes, glucose metabolism, and body composition. Diabetes 56(12):3020–3026. doi:10.2337/Db07-0338
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21(2):263–265. doi:10.1093/bioinformatics/bth457
Berthier M-T, Houde A, Paradis A-M, Couture P, Gaudet D, Despres J-P, Vohl M-C (2004) Molecular screening of the microsomal triglyceride transfer protein: association between polymorphisms and both abdominal obesity and plasma apolipoprotein B concentration. J Hum Genet 49(12):684–690
Bohme M, Grallert H, Fischer A, Gieger C, Nitz I, Heid I, Kohl C, Wichmann HE, Illig T, Doring F (2008) MTTP variants and body mass index, waist circumference and serum cholesterol level: association analyses in 7,582 participants of the KORA study cohort. Mol Genet Metab 95(4):229–232. doi:10.1016/j.ymgme.2008.09.004
Brouillette C, Bosse Y, Perusse L, Gaudet D, Vohl MC (2004) Effect of liver fatty acid binding protein (FABP) T94A missense mutation on plasma lipoprotein responsiveness to treatment with fenofibrate. J Hum Genet 49(8):424–432. doi:10.1007/s10038-004-0171-2
de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D (2005) Efficiency and power in genetic association studies. Nat Genet 37(11):1217–1223. doi:10.1038/ng1669
Donkor J, Zhang P, Wong S, O’Loughlin L, Dewald J, Kok BPC, Brindley DN, Reue K (2009) A conserved serine residue is required for the phosphatidate phosphatase activity but not the transcriptional coactivator functions of Lipin-1 and Lipin-2. J Biol Chem 284(43):29968–29978
Du H, van der AD, van Bakel MM, van der Kallen CJ, Blaak EE, van Greevenbroek MM, Jansen EH, Nijpels G, Stehouwer CD, Dekker JM, Feskens EJ (2008) Glycemic index and glycemic load in relation to food and nutrient intake and metabolic risk factors in a Dutch population. Am J Clin Nutr 87(3):655–661
Du H, Vimaleswaran KS, Angquist L, Hansen RD, van der AD, Holst C, Tjonneland A, Overvad K, Jakobsen MU, Boeing H, Meidtner K, Palli D, Masala G, Bouatia-Naji N, Saris WH, Feskens EJ, Wareham NJ, Sorensen TI, Loos RJ (2011) Genetic polymorphisms in the hypothalamic pathway in relation to subsequent weight change: the DiOGenes study. PLoS ONE 6(2):e17436. doi:10.1371/journal.pone.0017436
Ferguson PJ, Chen S, Tayeh MK, Ochoa L, Leal SM, Pelet A, Munnich A, Lyonnet S, Majeed HA, El-Shanti H (2005) Homozygous mutations in LPIN2 are responsible for the syndrome of chronic recurrent multifocal osteomyelitis and congenital dyserythropoietic anaemia (Majeed syndrome). J Med Genet 42(7):551–557. doi:10.1136/jmg.2005.030759
Fisher E, Weikert C, Klapper M, Lindner I, Möhlig M, Spranger J, Boeing H, Schrezenmeir J, Döring F (2007) L-FABP T94A is associated with fasting triglycerides and LDL-cholesterol in women. Mol Gen and Metabol 91:278–284
Fisher E, Meidtner K, Angquist L, Holst C, Hansen RD, Halkjaer J, Masala G, Ostergaard JN, Overvad K, Palli D, Vimaleswaran KS, Tjonneland A, van der AD, Wareham NJ, Sorensen T, Loos RJ, Boeing H (2012) Influence of dietary protein intake and glycemic index on the association between TCF7L2 HapA and weight gain. Am J Clin Nutr 95(6):1468–1476. doi:10.3945/ajcn.111.014670
Fougeray S, Loriot MA, Nicaud V, Legendre C, Thervet E, Pallet N (2011) Increased body mass index after kidney transplantation in activating transcription factor 6 single polymorphism gene carriers. Transplant Proc 43(9):3418–3422. doi:10.1016/j.transproceed.2011.09.022
Gauderman W, Morrison J (2006) QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies. http://hydra.usc.edu/gxe
Gordon DA, Jamil H (2000) Progress towards understanding the role of microsomal triglyceride transfer protein in apolipoprotein-B lipoprotein assembly. Biochim Biophys Acta 1486(1):72–83
Gropler MC, Harris TE, Hall AM, Wolins NE, Gross RW, Han X, Chen Z, Finck BN (2009) Lipin 2 is a liver-enriched phosphatidate phosphohydrolase enzyme that is dynamically regulated by fasting and obesity in mice. J Biol Chem 284(11):6763–6772. doi:10.1074/jbc.M807882200
Guillou H, Martin PG, Pineau T (2008) Transcriptional regulation of hepatic fatty acid metabolism. Subcell Biochem 49:3–47. doi:10.1007/978-1-4020-8831-5_1
He X, Xu X, Liu B (2009) Molecular characterization, chromosomal localization and association analysis with back-fat thickness of porcine LPIN2 and LPIN3. Mol Biol Rep 36(7):1819–1824. doi:10.1007/s11033-008-9385-2
Hussain MM, Shi J, Dreizen P (2003) Microsomal triglyceride transfer protein and its role in apoB-lipoprotein assembly. J Lipid Res 44(1):22–32
Hussain MM, Nijstad N, Franceschini L (2011) Regulation of microsomal triglyceride transfer protein. Clin Lipidol 6(3):293–303. doi:10.2217/clp.11.21
Iizuka K, Horikawa Y (2008) ChREBP: a glucose-activated transcription factor involved in the development of metabolic syndrome. Endocr J 55(4):617–624
Jäger R, Bertrand MJM, Gorman AM, Vandenabeele P, Samali A (2012) The unfolded protein response at the crossroads of cellular life and death during endoplasmic reticulum stress. Biol Cell 104(5):259–270. doi:10.1111/boc.201100055
Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI (2008) SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24(24):2938–2939. doi:10.1093/bioinformatics/btn564
Karpe F, Lundahl B, Ehrenborg E, Eriksson P, Hamsten A (1998) A common functional polymorphism in the promoter region of the microsomal triglyceride transfer protein gene influences plasma LDL levels. Arterioscler Thromb Vasc Biol 18(5):756–761
Kooner JS, Chambers JC, Aguilar-Salinas CA, Hinds DA, Hyde CL, Warnes GR, Gomez Perez FJ, Frazer KA, Elliott P, Scott J, Milos PM, Cox DR, Thompson JF (2008) Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat Genet 40(2):149–151. doi:10.1038/ng.2007.61
Larsen LH, Ängquist L, Vimaleswaran KS, Hager J, Viguerie N, Loos RJ, Handjieva-Darlenska T, Jebb SA, Kunesova M, Larsen TM, Martinez JA, Papadaki A, Pfeiffer AF, van Baak MA, Sorensen TI, Holst C, Langin D, Astrup A, Saris WH (2012) Analyses of single nucleotide polymorphisms in selected nutrient-sensitive genes in weight-regain prevention: the DIOGENES study. AJCN 95(5):1254–1260. doi:10.3945/ajcn.111.016543
Ledmyr H, Karpe F, Lundahl B, McKinnon M, Skoglund-Andersson C, Ehrenborg E (2002) Variants of the microsomal triglyceride transfer protein gene are associated with plasma cholesterol levels and body mass index. J Lipid Res 43(1):51–58
Ledmyr H, McMahon AD, Ehrenborg E, Nielsen LB, Neville M, Lithell H, MacFarlane PW, Packard CJ, Karpe F (2004) The microsomal triglyceride transfer protein gene-493T variant lowers cholesterol but increases the risk of coronary heart disease. Circulation 109(19):2279–2284. doi:10.1161/01.CIR.0000130070.96758.7b
Ledmyr H, Ottosson L, Sunnerhagen M, Ehrenborg E (2006) The Ile128Thr polymorphism influences stability and ligand binding properties of the microsomal triglyceride transfer protein. J Lipid Res 47(7):1378–1385. doi:10.1194/jlr.M600072-JLR200
Low S, Chin MC, Deurenberg-Yap M (2009) Review on epidemic of obesity. Ann Acad Med Singapore 38(1):57–59
Mansego ML, Martinez F, Martinez-Larrad MT, Zabena C, Rojo G, Morcillo S, Soriguer F, Martin-Escudero JC, Serrano-Rios M, Redon J, Chaves FJ (2012) Common variants of the liver fatty acid binding protein gene influence the risk of type 2 diabetes and insulin resistance in Spanish population. PLoS ONE 7(3):e31853. doi:10.1371/journal.pone.0031853
McArthur MJ, Atshaves BP, Frolov A, Foxworth WD, Kier AB, Schroeder F (1999) Cellular uptake and intracellular trafficking of long chain fatty acids. J Lipid Res 40(8):1371–1383
Meex SJ, van Greevenbroek MM, Ayoubi TA, Vlietinck R, van Vliet-Ostaptchouk JV, Hofker MH, Vermeulen VM, Schalkwijk CG, Feskens EJ, Boer JM, Stehouwer CD, van der Kallen CJ, de Bruin TW (2007) Activating transcription factor 6 polymorphisms and haplotypes are associated with impaired glucose homeostasis and type 2 diabetes in Dutch caucasians. J Clin Endocrinol Metab 92(7):2720–2725. doi:10.1210/jc.2006-2280
Meex SJR, Weissglas-Volkov D, van der Kallen CJH, Thuerauf DJ, van Greevenbroek MMJ, Schalkwijk CG, Stehouwer CDA, Feskens EJM, Heldens L, Ayoubi TA, Hofker MH, Wouters BG, Vlietinck R, Sinsheimer JS, Taskinen MR, Kuusisto J, Laakso M, de Bruin TWA, Pajukanta P, Glembotski CC (2009) The ATF6-Met[67] val substitution is associated with increased plasma cholesterol levels. Arterioscl Throm Vas 29(9):1322–1327. doi:10.1161/Atvbaha.108.180240
Newberry EP, Xie Y, Kennedy SM, Luo J, Davidson NO (2006) Protection against western diet-induced obesity and hepatic steatosis in liver fatty acid-binding protein knockout mice. Hepatology 44(5):1191–1205. doi:10.1002/hep.21369
Park JY, Mitrou PN, Keogh RH, Luben RN, Wareham NJ, Khaw KT (2011) Effects of body size and sociodemographic characteristics on differences between self-reported and measured anthropometric data in middle-aged men and women: the EPIC-Norfolk study. Eur J Clin Nutr 65(3):357–367. doi:10.1038/ejcn.2010.259
Parker A, Meyer J, Lewitzky S, Rennich JS, Chan G, Thomas JD, Orho-Melander M, Lehtovirta M, Forsblom C, Hyrkko A, Carlsson M, Lindgren C, Groop LC (2001) A gene conferring susceptibility to type 2 diabetes in conjunction with obesity is located on chromosome 18p11. Diabetes 50(3):675–680
Perneger TV (1998) What’s wrong with Bonferroni adjustments. BMJ 316(7139):1236–1238. doi:10.1136/bmj.316.7139.1236
Poupeau A, Postic C (2011) Cross-regulation of hepatic glucose metabolism via ChREBP and nuclear receptors. Biochim Biophys Acta 1812(8):995–1006. doi:10.1016/j.bbadis.2011.03.015
Randall JC, Winkler TW, Kutalik Z, Berndt SI, Jackson AU, Monda KL, Kilpeläinen TO, Esko T, Mägi R, Li S, Workalemahu T, Feitosa MF, Croteau-Chonka DC, Day FR, Fall T, Ferreira T, Gustafsson S, Locke AE, Mathieson I, Scherag A, Vedantam S, Wood AR, Liang L, Steinthorsdottir V, Thorleifsson G, Dermitzakis ET, Dimas AS, Karpe F, Min JL, Nicholson G, Clegg DJ, Person T, Krohn JP, Bauer S, Buechler C, Eisinger K, Bonnefond A, Froguel P, Hottenga J-J, Prokopenko I, Waite LL, Harris TB, Smith AV, Shuldiner AR, McArdle WL, Caulfield MJ, Munroe PB, Grönberg H, Chen Y-DI, Li G, Beckmann JS, Johnson T, Thorsteinsdottir U, Teder-Laving M, Khaw K-T, Wareham NJ, Zhao JH, Amin N, Oostra BA, Kraja AT, Province MA, Cupples LA, Heard-Costa NL, Kaprio J, Ripatti S, Surakka I, Collins FS, Saramies J, Tuomilehto J, Jula A, Salomaa V, Erdmann J, Hengstenberg C, Loley C, Schunkert H, Lamina C, Wichmann HE, Albrecht E, Gieger C, Hicks AA, Johansson Å, Pramstaller PP, Kathiresan S, Speliotes EK, Penninx B, Hartikainen A-L, Jarvelin M-R, Gyllensten U, Boomsma DI, Campbell H, Wilson JF, Chanock SJ, Farrall M, Goel A, Medina-Gomez C, Rivadeneira F, Estrada K, Uitterlinden AG, Hofman A, Zillikens MC, den Heijer M, Kiemeney LA, Maschio A, Hall P, Tyrer J, Teumer A, Völzke H, Kovacs P, Tönjes A, Mangino M, Spector TD, Hayward C, Rudan I, Hall AS, Samani NJ, Attwood AP, Sambrook JG, Hung J, Palmer LJ, Lokki M-L, Sinisalo J, Boucher G, Huikuri H, Lorentzon M, Ohlsson C, Eklund N, Eriksson JG, Barlassina C, Rivolta C, Nolte IM, Snieder H, Van der Klauw MM, Van Vliet-Ostaptchouk JV, Gejman PV, Shi J, Jacobs KB, Wang Z, Bakker SJL, Mateo Leach I, Navis G, van der Harst P, Martin NG, Medland SE, Montgomery GW, Yang J, Chasman DI, Ridker PM, Rose LM, Lehtimäki T, Raitakari O, Absher D, Iribarren C, Basart H, Hovingh KG, Hyppönen E, Power C, Anderson D, Beilby JP, Hui J, Jolley J, Sager H, Bornstein SR, Schwarz PEH, Kristiansson K, Perola M, Lindström J, Swift AJ, Uusitupa M, Atalay M, Lakka TA, Rauramaa R, Bolton JL, Fowkes G, Fraser RM, Price JF, Fischer K, KrjutÅ¡kov K, Metspalu A, Mihailov E, Langenberg C, Luan Ja, Ong KK, Chines PS, Keinanen-Kiukaanniemi SM, Saaristo TE, Edkins S, Franks PW, Hallmans G, Shungin D, Morris AD, Palmer CNA, Erbel R, Moebus S, Nöthen MM, Pechlivanis S, Hveem K, Narisu N, Hamsten A, Humphries SE, Strawbridge RJ, Tremoli E, Grallert H, Thorand B, Illig T, Koenig W, Müller-Nurasyid M, Peters A, Boehm BO, Kleber ME, März W, Winkelmann BR, Kuusisto J, Laakso M, Arveiler D, Cesana G, Kuulasmaa K, Virtamo J, Yarnell JWG, Kuh D, Wong A, Lind L, de Faire U, Gigante B, Magnusson PKE, Pedersen NL, Dedoussis G, Dimitriou M, Kolovou G, Kanoni S, Stirrups K, Bonnycastle LL, Njølstad I, Wilsgaard T, Ganna A, Rehnberg E, Hingorani A, Kivimaki M, Kumari M, Assimes TL, Barroso I, Boehnke M, Borecki IB, Deloukas P, Fox CS, Frayling T, Groop LC, Haritunians T, Hunter D, Ingelsson E, Kaplan R, Mohlke KL, O’Connell JR, Schlessinger D, Strachan DP, Stefansson K, van Duijn CM, Abecasis GR, McCarthy MI, Hirschhorn JN, Qi L, Loos RJF, Lindgren CM, North KE, Heid IM, Consortium D, Investigators M (2013) Sex-stratified Genome-wide Association Studies Including 270,000 Individuals Show Sexual Dimorphism in Genetic Loci for Anthropometric Traits. PLoS Genet 9(6):e1003500. doi:10.1371/journal.pgen.1003500
Reue K, Donkor J (2007) Genetic factors in type 2 diabetes: all in the (lipin) family. Diabetes 56(12):2842–2843. doi:10.2337/db07-1288
Reue K, Dwyer JR (2009) Lipin proteins and metabolic homeostasis. J Lipid Res 50(Suppl (Supplement)):S109–S114. doi:10.1194/jlr.R800052-JLR200
Riboli E, Kaaks R (1997) The epic project: rationale and study design. European prospective investigation into cancer and nutrition. Int J Epidemiol 26(Suppl 1):S6–S14
Riboli E, Hunt KJ, Slimani N, Ferrari P, Norat T, Fahey M, Charrondiere UR, Hemon B, Casagrande C, Vignat J, Overvad K, Tjonneland A, Clavel-Chapelon F, Thiebaut A, Wahrendorf J, Boeing H, Trichopoulos D, Trichopoulou A, Vineis P, Palli D, Bueno-De-Mesquita HB, Peeters PH, Lund E, Engeset D, Gonzalez CA, Barricarte A, Berglund G, Hallmans G, Day NE, Key TJ, Kaaks R, Saracci R (2002) European prospective investigation into cancer and nutrition (EPIC): study populations and data collection. Public Health Nutr 5(6B):1113–1124. doi:10.1079/PHN2002394
Rinaldi S, Key TJ, Peeters PH, Lahmann PH, Lukanova A, Dossus L, Biessy C, Vineis P, Sacerdote C, Berrino F, Panico S, Tumino R, Palli D, Nagel G, Linseisen J, Boeing H, Roddam A, Bingham S, Khaw KT, Chloptios J, Trichopoulou A, Trichopoulos D, Tehard B, Clavel-Chapelon F, Gonzalez CA, Larranaga N, Barricarte A, Quiros JR, Chirlaque MD, Martinez C, Monninkhof E, Grobbee DE, Bueno-de-Mesquita HB, Ferrari P, Slimani N, Riboli E, Kaaks R (2006) Anthropometric measures, endogenous sex steroids and breast cancer risk in postmenopausal women: a study within the EPIC cohort. Int J Cancer 118(11):2832–2839. doi:10.1002/ijc.21730
Robitaille J, Brouillette C, Lemieux S, Perusse L, Gaudet D, Vohl MC (2004) Plasma concentrations of apolipoprotein B are modulated by a gene–diet interaction effect between the LFABP T94A polymorphism and dietary fat intake in French–Canadian men. Mol Genet Metab 82(4):296–303. doi:10.1016/j.ymgme.2004.06.002
Rubin D, Helwig U, Pfeuffer M, Schreiber S, Boeing H, Fisher E, Pfeiffer A, Freitag-Wolf S, Foelsch UR, Doering F, Schrezenmeir J (2006) A common functional exon polymorphism in the microsomal triglyceride transfer protein gene is associated with type 2 diabetes, impaired glucose metabolism and insulin levels. J Hum Genet 51(6):567–574. doi:10.1007/s10038-006-0400-y
Rubin D, Schneider-Muntau A, Klapper M, Nitz I, Helwig U, Folsch UR, Schrezenmeir J, Doring F (2008) Functional analysis of promoter variants in the microsomal triglyceride transfer protein (MTTP) gene. Hum Mutat 29(1):123–129. doi:10.1002/humu.20615
R Development Core Team (2010) R: a language and environment for statistical computing. Version 2.11.1, http://www.R-project.org edn. R Foundation for Statistical Computing, Vienna, Austria
Salminen A, Kaarniranta K (2010) ER stress and hormetic regulation of the aging process. Ageing Res Rev 9(3):211–217. doi:10.1016/j.arr.2010.04.003
Saris WH, Harper A (2005) DiOGenes: a multidisciplinary offensive focused on the obesity epidemic. Obes Rev 6(2):175–176
Schwarzer G (2010) Meta: meta-analysis with R. R package version 1.6-0; http://CRAN.R-project.org/package=meta edn
St-Pierre J, Lemieux I, Miller-Felix I, Prud’homme D, Bergeron J, Gaudet D, Nadeau A, Despres JP, Vohl MC (2002) Visceral obesity and hyperinsulinemia modulate the impact of the microsomal triglyceride transfer protein −493G/T polymorphism on plasma lipoprotein levels in men. Atherosclerosis 160(2):317–324
Talmud PJ, Drenos F, Shah S, Shah T, Palmen J, Verzilli C, Gaunt TR, Pallas J, Lovering R, Li K, Casas JP, Sofat R, Kumari M, Rodriguez S, Johnson T, Newhouse SJ, Dominiczak A, Samani NJ, Caulfield M, Sever P, Stanton A, Shields DC, Padmanabhan S, Melander O, Hastie C, Delles C, Ebrahim S, Marmot MG, Smith GD, Lawlor DA, Munroe PB, Day IN, Kivimaki M, Whittaker J, Humphries SE, Hingorani AD (2009) Gene-centric association signals for lipids and apolipoproteins identified via the human CVD beadchip. Am J Hum Genet 85(5):628–642. doi:10.1016/j.ajhg.2009.10.014
Thameem F, Farook VS, Bogardus C, Prochazka M (2006) Association of amino acid variants in the activating transcription factor 6 gene (ATF6) on 1q21-q23 with type 2 diabetes in Pima Indians. Diabetes 55(3):839–842
The International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437(7063):1299–1320
van Bakel MM, Kaaks R, Feskens EJ, Rohrmann S, Welch AA, Pala V, Avloniti K, van der Schouw YT, Van der AD, Du H, Halkjaer J, Tormo MJ, Cust AE, Brighenti F, Beulens JW, Ferrari P, Biessy C, Lentjes M, Spencer EA, Panico S, Masala G, Bueno-de-Mesquita HB, Peeters PH, Trichopoulou A, Psaltopoulou T, Clavel-Chapelon F, Touvier M, Skeie G, Rinaldi S, Sonestedt E, Johansson I, Schulze M, Ardanaz E, Buckland G, Tjonneland A, Overvad K, Bingham S, Riboli E, Slimani N (2009) Dietary glycaemic index and glycaemic load in the European Prospective Investigation into Cancer and Nutrition. Eur J Clin Nutr 63(Suppl 4):S188–S205. doi:10.1038/ejcn.2009.81
Veerkamp JH, van Moerkerk HT (1993) Fatty acid-binding protein and its relation to fatty acid oxidation. Mol Cell Biochem 123(1–2):101–106
Vimaleswaran KS, Angquist L, Hansen RD, van der A DL, Bouatia-Naji N, Holst C, Tjonneland A, Overvad K, Jakobsen MU, Boeing H, Meidtner K, Palli D, Masala G, Saris WHM, Feskens EJM, Wareham NJ, Sorensen TIA, Loos RJF (2012) Association Between FTO Variant and Change in Body Weight and Its Interaction With Dietary Factors: The DiOGenes Study. Obesity 20 (8):1669–1674. doi: 10.1038/Oby.2012.49
von Ruesten A, Steffen A, Floegel A, van der A DL, Masala G, Tjønneland A, Halkjaer J, Palli D, Wareham NJ, Loos RJF, Sørensen TIA, Boeing H (2011) Trend in Obesity Prevalence in European Adult Cohort Populations during Follow-up since 1996 and Their Predictions to 2015. PLoS ONE 6 (11):e27455. doi:10.1371/journal.pone.0027455
Weickert MO, Cv L, Roden M, Chandramouli V, Brehm A, Nowotny P, Osterhoff MA, Isken F, Spranger J, Landau BR, Pfeiffer AFH, Möhlig M (2007) A Thr94Ala mutation in human liver fatty acid-binding protein contributes to reduced hepatic glycogenolysis and blunted elevation of plasma glucose levels in lipid-exposed subjects. Am J Physiol Endocrinol Metab 293(4):E1078–E1084. doi:10.1152/ajpendo.0 0337.2007
WHO (2000) Obesity: preventing and managing the global epidemic. Report of a WHO consultation. Report No: 894. WHO, Geneva
Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR (2008) Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 40(2):161–169. doi:10.1038/ng.76
Zeng L, Lu M, Mori K, Luo S, Lee AS, Zhu Y, Shyy JY (2004) ATF6 modulates SREBP2-mediated lipogenesis. EMBO J 23(4):950–958. doi:10.1038/sj.emboj.7600106
Acknowledgments
The European Prospective Investigation into Cancer and Nutrition (EPIC) (http://epic.iarc.fr/) generously contributed with its data. The present study was supported by the integrative DiOGenes project. DiOGenes is the acronym of the project’ Diet, Obesity and Genes’ supported by the European Community (Contract no. FOOD-CT-2005-513946). We also would like to acknowledge the support of the Federal Ministry of Education and Research for research into determinants of weight gain in Germany (FKZ: 01ER0808).
Ethical standards
The Sponsors of the DiOGenes Project are listed on the website of the project (http://www.diogenes-eu.org/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Karina Meidtner, Eva Fisher, Lars Ängquist, Claus Holst, Karani S Vimaleswaran, Jolanda MA Boer, Jytte Halkjær, Giovanna Masala, Jane N Østergaard, Lotte M Mortensen, Daphne L. van der A, Anne Tjønneland, Domenico Palli, Kim Overvad, Nicholas J Wareham, Ruth JF Loos, Thorkild IA Sørensen and Heiner Boeing declare that they have no conflict of interest. All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000. Informed consent was obtained from all patients for being included in the study.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Meidtner, K., Fisher, E., Ängquist, L. et al. Variation in genes related to hepatic lipid metabolism and changes in waist circumference and body weight. Genes Nutr 9, 385 (2014). https://doi.org/10.1007/s12263-014-0385-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12263-014-0385-7