Abstract
Switchgrass (Panicum virgatum L.) is a native perennial warm season (C4) grass that has been identified as a promising species for bioenergy research and production. Consequently, biomass yield and feedstock quality improvements are high priorities for switchgrass research. The objective of this study was to develop a switchgrass genetic linkage map using a full-sib pseudo-testcross mapping population derived from a cross between two heterozygous genotypes selected from the lowland cultivar ‘Alamo’ (AP13) and the upland cultivar ‘Summer’ (VS16). The female parent (AP13) map consists of 515 loci in 18 linkage groups (LGs) and spans 1,733 cM. The male parent (VS16) map arranges 363 loci in 17 LGs and spans 1,508 cM. No obvious cause for the lack of one LG in VS16 could be identified. Comparative analyses between the AP13 and VS16 maps showed that the two major ecotypic classes of switchgrass have highly colinear maps with similar recombination rates, suggesting that chromosomal exchange between the two ecotypes should be able to occur freely. The AP13 and VS16 maps are also highly similar with respect to marker orders and recombination levels to previously published switchgrass maps. The genetic maps will be used to identify quantitative trait loci associated with biomass and quality traits. The AP13 genotype was used for the whole genome-sequencing project and the map will thus also provide a tool for the anchoring of the switchgrass genome assembly.
Similar content being viewed by others
Introduction
Switchgrass (Panicum virgatum L.) is a perennial warm season (C4) grass native to North America. The species is largely self-incompatible and, consequently, mainly outcrossing and highly heterozygous. It has been used as a forage crop and for soil conservation, and was identified in the 1990s as a potential feedstock crop for the production of cellulosic biofuels [1, 2]. High biomass yield (high net energy production per unit area), broad adaptability including in marginal areas, low production costs, low nutrient requirements, and high water use efficiency [1, 3, 4] are some of the beneficial attributes that favor switchgrass for bioenergy production.
The species is categorized into two major ecotypes based on plant morphology [5] and adaptation area [6]. The lowland types are tall, coarse in leaf texture, and adapted to the flood plains. Upland types are shorter, have finer leaves, and a slower growth rate than the lowland types and are adapted to the northern USA [1, 4]. Lowland ecotypes are primarily tetraploid (2n = 4x = 36) while upland ecotypes can be tetraploid but are predominantly octoploid (2n = 8x = 72) [7–9]. Although lowland switchgrass had originally been considered an autotetraploid with a high degree of preferential pairing [10], genetic mapping has strongly suggested that it is a disomic tetraploid [11]. The recent observation that the E genome of species belonging to section Rudgeani within the genus Panicum is not phylogenetically equidistant to both switchgrass subgenomes also supports that switchgrass is an allopolyploid [12].
Biomass yield improvement and the reduction of recalcitrance, which describes the unavailability of sugars for fermentation, of switchgrass through breeding and genetic manipulation are priority research areas of the Bioenergy Science Center (BESC), one of three DOE-funded Bioenergy Research Centers. The availability of genetic variability for the traits of interest, their heritability, the selection intensity on those traits, and the efficiency of the breeding procedure are key elements for the success of a breeding program [13]. The development of a genetic map to dissect traits into their genetic components and determine the location of those components is a first step towards enhancing the breeding process to maximize biomass yield and optimize the cell wall composition for bioenergy production through fermentation.
Three genetic maps have been produced in switchgrass [10, 11, 14]. Missaoui et al. [10] used restriction fragment length polymorphism (RFLP) markers in a full-sib progeny population from a cross between a lowland genotype derived from the cultivar Alamo (AP13) and an upland genotype derived from the cultivar Summer (VS16). Due to the relatively small number of mapped markers and the small size of the mapping population (85 progeny), genome coverage and mapping power were low. Okada et al. [11] constructed a switchgrass map using a full-sib pseudo-testcross mapping population derived from a cross between two lowland genotypes. Single-dose microsatellite markers were mapped to generate female and male maps which spanned 1,376 and 1,645 cM, respectively. Recently, Liu and colleagues generated a genetic map in a population obtained by selfing a self-compatible lowland genotype NL94 LYE [14]. The maps comprised 499 SSR loci and spanned 2,085 cM. A complete map of switchgrass which includes both lowland and upland ecotypes remains of prime importance.
The objectives of this research were to (1) increase the density and resolution of the AP13 × VS16 map begun by Missaoui et al. [10] using SSR and diversity array technology (DArT) markers and a larger population; (2) study the inheritance of the markers in the two ecotypes; and (3) conduct a comparative analysis of the resulting maps with the lowland × lowland maps generated by Okada et al. [11].
Materials and Methods
Mapping Population
The mapping population was developed by crossing the lowland genotype ‘AP13’, derived from cv. ‘Alamo’ as the female parent with an upland genotype ‘VS16’, derived from cv. ‘Summer’ as the male parent. Both genotypes are tetraploid with an expected somatic chromosome number of 2n = 4x = 36. The salient features of AP13 are high biomass yield, early and vigorous growth, late maturity, and moderate resistance to rust (Puccinia emaculata Schwein). In contrast, VS16 is late in spring regrowth, less vigorous, highly susceptible to rust, early maturing, shorter in stature, and low in biomass yield. A first full-sib population between the AP13 and VS16 clones, consisting of 85 individuals, was developed at the University of Georgia and used by Missaoui et al. [10]. To increase the population size, additional crosses were made in 2006 between the same clonally propagated parents at The Samuel Roberts Noble Foundation (NF), Ardmore, OK. The expanded population consisted of 191 F1 plants, including 62 plants previously included in the Missaoui map [10].
DNA Extraction
Total genomic DNA was extracted from young leaves. Fresh leaves of the parents and each of the 191 progeny were harvested from a clonal plant copy kept in the greenhouse and immediately frozen in liquid nitrogen. The tissue was disrupted and homogenized with the TissueLyser, and DNA was extracted using a slightly modified DNeasy Plant Mini Kit protocol (QIAGEN Inc., Valencia, CA), which involved adding 550 μl AP1 (preheated to 65 °C) cell lysis buffer, 5 μl of a 100 mg/ml RNase A stock, 182 μl AP2 buffer, and 743 μl AP3/E to each tube of ground tissue, washing with 600 μl AW buffer and elution with 165 μl AE buffer. DNA concentrations were measured using a NanoDrop, ND 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and DNA quality and integrity were assessed by electrophoresis on 1.0 % (w/v) agarose gels stained with ethidium bromide.
Marker Development
SSR Markers
Genomic SSR markers were isolated from switchgrass using two different protocols. At the Noble Foundation, a (GA/CT) n -enriched genomic library was constructed from switchgrass genotype AP13 using the FIASCO protocol [15]. Primer sets were designed from sequences containing SSRs of at least 18 bp (NFSG markers). At the University of Georgia (UGA), di- and trinucleotide SSRs were isolated from PstI 1.2–2.0 kb fragment libraries, also of genotype AP13, following hybridization of 92,160 colonies with the SSR oligo probes (GA)15, (CA)15, (GGA)10, and (GCA)10 as described by Dida et al. [16]. A subset of the clones was also screened with (ACC)10 and (GAC)10 (55,296 clones), and with (GAA)10 (18,432 clones). Clones with strong hybridization signals were sequenced using Sanger technology. Primers were designed against sequences containing SSRs of at least 10 bp in length for mononucleotide repeats, 16 bp for dinucleotide repeats (8 repeat units) and 21 bp for trinucleotide repeats (7 repeat units; UGSW markers). All primer design was carried out using the software Primer 3 [17].
In addition to 1,248 genomic SSR primer pairs (NFSG and UGSW markers), 349 EST-SSR primer pairs (SWW markers) [18, 19] were tested on the two parents and six randomly selected individuals from the mapping population to determine marker amplification and ability to reveal polymorphism. Primer sequences used for mapping are listed in Electronic Supplementary Material (ESM) 1 (UGSW and newly developed NFSG markers) or had previously been published [11].
EST-STS Markers
A total of 144 primer pairs was generated from cell-wall-related gene sequences. The gene sequences for different subunits of cellulose synthases, transferases, decarboxylases, and epimerases from maize, sorghum, rice, and Arabidopsis found in the NCBI unigene database (http://www.ncbi.nlm.nih.gov;Unigene) were used as queries in BLASTN searches against the switchgrass EST database (www.switchgrassgenomics.noble.org). Primers were developed to the ESTs with the highest identity and lowest E-value including 73 cellulose synthases, four alpha-1,4-glucansynthases, three 1,4-beta-d-glucan synthases, six starch synthases, eight glucomannon 4-betatransferases, six putative xyloglucan glycosyltransferases, six probable mannonsynthases, five glucose-1-phosphate adenylyltransferases, one each of UDP glucuronate decarboxylase, UDP xylose synthase, and UDP-d-mannose 3′,5′-epimerase, and 30 unitranscripts with unknown function. EST-STS markers were given the prefix NFSTS. Primer sequences for NFSTS markers placed on the genetic map are listed in ESM 1.
Diversity Array Technology Markers
The microarray-based DArT markers were developed by first testing eight combinations of the rare-cutting restriction enzyme PstI with different restriction endonucleases that cut frequently on DNA samples from AP13 and VS16 in order to identify the combination that resulted in the most heterodispersed smear of restriction fragments (absence of any noticeable bands). The combination of PstI and MspI was selected to construct two libraries of 6,144 and 1,536 genomic clones (7,680 clones in total) as described by Jaccoud et al. [20]. In order to produce genomic representations, approximately 50 ng of genomic DNA was digested with PstI/MspI combinations and the resulting fragments ligated to a PstI overhang compatible oligonucleotide adapter. A primer annealing to this adapter was used in a PCR reaction to amplify genomic fragments which were cloned into the pCR2.1-TOPO vector (Invitrogen, Australia) as described by Jaccoud et al. [20]. The white colonies containing switchgrass genomic fragments were picked into individual wells of 384-well microtiter plates filled with ampicillin/kanamycin-supplemented freezing medium [21]. Inserts from these clones were amplified using M13F and M13R primers in 384 plate format. PCR products were dried, washed, and dissolved in a spotting buffer. The amplification products were used as probes for printing DArT arrays. DArT arrays were printed on SuperChip poly-L-lysine slides (Thermo Scientific) using a MicroGrid arrayer (Genomics Solutions) using 7,680 inserts (all printed in replication).
Each sample (parents and 135 progenies) was assayed using methods described above for library construction. Genomic representations were labeled with fluorescent dyes (Cy3 and Cy5). Labeled targets were then hybridized to printed DArT arrays for 16 h at 62 °C in a water bath. Slides were processed as described by Kilian et al. [21] and scanned using a Tecan LS300 scanner (Tecan Group Ltd, Männedorf, Switzerland) generating three images per array: one image scanned at 488 nm measures the amount of DNA within the spot based on the hybridization signal of a FAM-labeled fragment of a TOPO vector multiple cloning site fragment (reference signal) and two images for “target” signal measurement. Signal intensities were extracted from images using DArTsoft 7.4.7 software (http://www.diversityarrays.com/software.html). DArTsoft was also used to convert signal intensities to presence/absence (binary) scores used in the downstream analysis. Both DArT assays and DArtsoft analysis were performed at DArT PL in Canberra, Australia.
PCR Protocol and Genotyping
SWW and NFSG markers were generated as described [11]. UGSW markers were amplified in a total volume of 10 μl consisting of 50–100 ng genomic DNA, 200 nM tailed (21 bp M13 tail: 5′-CGTTGTAAAACGACGGCCAGT-3′) forward primer, 400 nM M13-labeled primer, 0.8 μM reverse primer, 0.8 U GoTaq DNA Polymerase (Promega, Madison, WI), 3 mM MgCl2, 0.25 mM dNTPs, 0.13 mM DDT, 1.3 % DMSO, and 0.54 mM betaine in 1X buffer. PCR consisted of an initial denaturation of 3 min at 94 °C, three touchdown cycles of denaturation at 94 °C for 30 s, annealing at 68 °C with a decrease of 2 °C/cycle for 1 min and extension at 72 °C for 1 min, and 32 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s and extension at 72 °C for 45 s. The final extension was held at 72 °C for 10 min after which the samples were cooled to 4 °C.
Amplification products from three or four primer sets labeled with different fluorochromes were pooled and 2–5 μl of the pooled amplicons were added to 8–10 μl Hi-Di formamide (Life Technologies Corporation, Carlsbad, CA) and, depending on the fluorochromes used, 0.2 ul of LIZ500 or 0.5 ul of ROX size standard (Life Technologies Corporation, Carlsbad, CA). Multiplexed PCR products were analyzed on an ABI 3730xl sequencer and results were scored using GeneMapper v. 3.7 or v 4.0 (Life Technologies Corporation, Carlsbad, CA).
Marker Segregation and Linkage Analysis
Switchgrass is an outcrossing tetraploid and, hence, as many as four alleles of a single copy gene may be PCR-amplified using a single primer set in a given individual. To facilitate the scoring of the SSR and EST-STS markers, multiple fragments amplified from a single primer set and differentially present in the parents were considered independent markers. The segregation of each fragment was tested for fit to a 1:1 ratio using a chi-square test. Initially, only markers segregating in a 1:1 ratio were used for map construction because the inclusion of distorted markers generated many spurious linkages. For DArT markers, in addition to a 1:1 segregation, a marker P value >70 was required for inclusion in the mapping data set. All dominantly scored marker data, irrespective of the parent of origin, were jointly analyzed using the software package JoinMap 4.0 [22] with the double pseudo-testcross strategy [23] and the doubled haploid model. Calculation of the linkage maps was done using the regression mapping algorithm at a pairwise recombination frequency estimate <0.40, a logarithm (base 10) of odds (LOD) score ≥3, a goodness-of-fit jump threshold of 5, and a ripple value of 1. The Kosambi mapping function was used to convert recombination units into genetic distances [24]. LGs that consisted of fragments that were present in AP13 were identified as female and LG with fragments that were present in VS16 were identified as male.
To enhance the quality of the genetic maps, DArT markers with a mean chi-square contribution >2.5 were removed if they had P values <80. SSR and EST-STS markers with a high mean chi-square contribution were rescored and, if the chi-square contribution remained high, sequentially removed until all markers had a chi-square contribution ≤3.0. Linkage groups with ≤3 markers and LGs containing ≥50 % of DArT markers with a P value <80 were not included in the final maps.
The marker set remaining after removal of low-quality markers was then used to generate genetic maps using the software program Mapmaker [25, 26]. The population type was set as a backcross, and LGs were obtained at a LOD score of 4 with a maximum distance of 35 cM. Marker orders were obtained using a combination of three-point and multipoint analyses and final orders were tested using the ‘ripple’ command. Double recombination events were ignored in the calculation of the genetic distances. Distances were given in Kosambi centiMorgans [24]. Mapmaker map orders were scrutinized manually for the presence of recombination events, and markers that could not be ordered unambiguously were indicated on the maps (Fig. 1). If, after considering ambiguous markers orders, discrepancies remained between the JoinMap and Mapmaker maps (ESM 2), we used the ‘compare’ function in Mapmaker to calculate the LOD score of the obtained JoinMap and Mapmaker orders relative to the best order for the discrepant region. If necessary, Mapmaker maps were adjusted to reflect the most likely order of markers. The LOD scores of discrepant marker orders in the JoinMap maps relative to the Mapmaker maps are given in ESM 2.
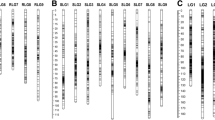
Genetic maps constructed using the software package Mapmaker. LG numbering is taken from Okada et al. [11]. Loci identified in homologous female (f) and male (m) maps are given in bold and connected by lines. Genetic distances are given in cM (Kosambi). Loci with distorted segregation ratios are indicated with * (0.01 < P ≤ 0.05), ** (0.001 < P ≤ 0.01), and *** (P ≤ 0.001). Vertical bars next to loci names indicate loci that could not be ordered unambiguously
To ensure that we did not miss chromosomes or chromosome regions by excluding markers that did not segregate in a 1:1 ratio, we conducted a two-point analysis on the entire data file (consisting of markers with 1:1 and distorted segregation ratios) and formed LGs at a series of increasing LOD scores. Groups of loci that remained together at increasing LOD scores were considered as putatively belonging to a single LG and were further analyzed by three-point and multipoint analyses. Highly distorted markers that were flanked on either side by a non-distorted marker were removed from the map as they typically generated many double recombination events suggesting that the distortion was due to scoring errors or the colocation of fragments.
Intraspecific Comparative Analysis
Markers that identified shared loci on the male and female maps were used to identify LGs that were likely homologous. Since switchgrass is a tetraploid, some primer pairs also amplified DNA fragments in both switchgrass subgenomes and these markers were used to identify homoeologous linkage groups. Two female (a and b) and two male LGs (a and b) that carried common markers were therefore allocated to the same homoeologous group. Comparative analyses were carried out between male and female maps, and between our maps and the maps previously published by Okada et al. [11]. In addition to considering conservation of marker orders, we also determined whether map lengths were similar. This was accomplished by conducting paired t tests on all common marker intervals in the entire map. We also totaled the length of the common intervals per LG, and conducted paired t tests on the totals.
Results
Type of Markers and Their Polymorphism Levels
Genomic SSR Markers
Screening of randomly selected 1.2–2.0 kb switchgrass PstI clones by hybridization with di- and trinucleotide repeats indicated that 1.15 % of the clones contained (GA) n repeats, 0.87 % contained (CA) n repeats, 0.71 % contained (GGA) n repeats, 0.53 % contained (GCA) n repeats, 0.55 % contained (ACC) n repeats, 0.49 % contained (GAC) n repeats, and 0.02 % contained (GAA) n repeats. Of the 3,591 positive clones identified, 2,304 clones with strong hybridization signals were selected for Sanger sequencing, yielding 2.8 Mb of sequence data for 2,269 clones. Sixty-nine percent of the sequenced clones contained at least five units of a dinucleotide or trinucleotide repeat. This number dropped to 40 % for a SSR length of at least 7 repeat units. A total of 736 primer pairs were designed against SSRs present in 683 clones, 96.7 % of which successfully amplified and 57 % of which segregated in six switchgrass genotypes consisting of the parents of the mapping population and four randomly selected progeny.
A second source of switchgrass genomic SSRs was a (GA/CT) n enriched library. A total of 4,992 clones was sequenced, yielding 41 % non-redundant sequences of which 26 % contained dinucleotide repeats with an SSR length of at least 6 repeat units. Of the 512 primer pairs designed, 67 % successfully amplified DNA from the parents and a subset of the progeny and 54 % revealed polymorphisms.
EST-SSR and EST-STS Markers
Eighty-nine (26 %) out of 349 EST-SSR primer pairs tested detected variation in the AP13 × VS16 mapping population. Of the 144 primer pairs developed against cell-wall-related genes, 78 % amplified well in switchgrass, but only 18 % were polymorphic in the mapping population.
DArT Markers
DArTsoft analysis of the parents and the progenies identified 633 segregating DArT markers using standard thresholds, which represents 8.2 % of the markers on the array (7,680 markers printed). The average call rate was at 95 % which is slightly lower than in a typical DArT analysis (97 %) and scoring discordance (measuring frequency of scoring errors) at 0.004 was slightly higher than normal (typically below 0.002).
Marker Segregation and Linkage Mapping
A set of 475 primer pairs was used to genotype the mapping population, generating a total of 947 scorable fragments that were present in one parent and absent (or presence unknown) in the other parent. Because all fragments amplified by a single primer were scored independently, several of these fragments were expected to be allelic; 80.3 % of the fragments segregated in a 1:1 ratio, 10.0 % had marginally distorted segregation ratios (0.01 < P ≤ 0.05), and 9.7 % had severely distorted segregation ratios (P ≤ 0.01). The latter category may include fragments that were present in both parents, but could only be scored in one of the parents. A breakdown of these percentages for the EST-SSR, genomic SSR, and EST-STS markers is given in Table 1. Only markers that fit a 1:1 segregation ratio were used in the construction of the initial linkage map.
In addition, a DArT array containing 7,680 clones was used to genotype a subset (135 progeny) of the mapping population. Segregation was obtained for 633 markers (8.2 %), 61 % of which segregated in a 1:1 ratio. P values, which indicate marker quality, varied from 50 to 97 %. For the initial mapping, we arbitrarily chose 70 % as a cut-off point for including markers in the analysis. This removed 6 % of DArT markers that segregated in a 1:1 ratio. We also removed DArT markers that hybridized to both parents and those that hybridized to neither parent. In total, 760 PCR fragments and 341 DArT markers were included in the dataset used for the construction of the initial linkage maps. Grouping of the data at a LOD score ≥3 identified 18 female LGs and 17 male LGs containing a minimum of four non-allelic markers. PCR-fragments generated by the same primer set that co-localized on the genetic map or for which the genotypic scores differed only by double recombination events were consolidated into a single locus.
Placement of markers with distorted segregation ratios onto the framework map showed that more than 60 % of the severely distorted markers (P < 0.01) did not define specific chromosome regions but were dispersed among the framework markers and/or generated many double cross-over events. This suggested that the distortion was caused by technical issues (e.g., cosegregation of fragments, scoring errors) rather than biological factors, and therefore these markers were not added to the map. However, three clusters of distorted markers were identified; a cluster of 18 markers formed a separate VS16 LG, a second cluster of seven markers joined two VS16 LGs, and a third cluster consisting of five markers extended a VS16 LG with 33.2 cM. The final dataset consisted of 772 non-distorted markers, 74 marginally distorted markers, and 32 severely distorted markers (ESM 3).
Using this final dataset, genetic maps were constructed with both JoinMap and Mapmaker. The two software programs yielded the same groupings, but some differences in marker orders were observed for 16 out of the 18 female LGs and eight out of the 17 male LGs (ESM 2). Surprisingly, in more than 75 % of the cases, the JoinMap order appeared to be at least 100 times less likely (LOD < −2.00) than the most likely Mapmaker order (ESM 2). All subsequent analyses were therefore conducted on the Mapmaker maps (Fig. 1).
The Organization of LGs into Homoeologous Groups
The female (AP13) map comprises 515 loci organized into 18 LGs and spans a total of 1,733 cM. The male (VS16) map consists of 363 loci organized in 17 LGs and spans 1,508 cM (Fig. 1). The length of individual LGs varies from 58 to 149 cM in the female map and from 52 to 151 cM in the male map. One hundred and six primer sets generated markers that could be mapped in both the male and female maps. Female and male LGs that contained a minimum of three common markers were aligned into homologous groups. Fifteen homologous groups were identified that each consisted of one female and one male LG. No female LG could be identified that were homologous to the two remaining male LGs. In addition to linking the AP13 and VS16 maps, markers that are heterozygous and have different alleles in both parents also provide an easy tool to check for selfed progeny within the pseudo F1 testcross population. All progeny carried both female and male alleles which demonstrates their hybrid origin.
Because our linkage map included 51 EST-SSR and 117 gSSR loci that had previously been mapped by Okada et al. [11], we could directly compare the two sets of maps. The 15 homologous groups corresponded to LGs I-a, I-b, II-b, III-a, III-b, IV-a, IV-b, V-a, V-b, VI-a, VI-b, VII-b, VIII-b, IX-a, and IX-b in Okada et al. [11], and were given the same designations. It should be noted that ‘A’ and ‘B’ designations used by Okada and colleagues [11] and thus also the corresponding ‘a’ and ‘b’ designations used here (lower case letters were used in this paper to avoid connotation to subgenomes) were assigned arbitrarily to pairs of homoeologous chromosomes and do not imply allocation of linkage groups to subgenomes. The three female LGs for which no male homolog was identified in our data set corresponded to LGs II-a, VII-a, and VIII-a. Revisiting the two male LGs for which we had not been able to identify a female homolog, one has two markers in common with LG VIII-b and the other has one marker in common with LG II-b presented in this paper. It seems therefore likely that they correspond to LGs VIII-a and II-a.
Intraspecific Comparative Analyses
Comparison of the Female and Male Maps
Marker orders in the male and female maps were completely colinear in all LGs except LG VII-b where the three common markers were in the order NFSG192—11.8 cM–UGSW86—13.4 cM–NFSG224 in the female map and NFSG192—33.2 cM–NFSG224—8.6 cM–UGSW86 in the male map. It is unclear whether this represents a rearrangement in the male map compared to the female map, or whether two independent loci were mapped for one of the three markers in the AP13 and VS16 maps. Using marker intervals that were common between the female and male LGs but excluding the VII-b intervals, we examined whether recombination was significantly different in the female and male maps. Although the overall length of the common intervals was some 7 % shorter in the male compared to the female maps, a paired t test showed that there was no significant length difference at either the interval level or at the LG level (P > 0.05).
Comparison with Previously Published Maps
A total of 73 primer sets detected loci that were colinear between homologous LGs in our AP13 maps and the Alamo maps published by Okada and colleagues [11], and the same number of markers detected common loci between homologous AP13 and Okada Kanlow LGs. The number of common markers between homologous LGs in the VS16, and the Okada Alamo and Kanlow maps was 43 and 44, respectively. The map length defined by the common marker intervals was not significantly different in the AP13–Alamo (703 cM vs. 774 cM; P = 0.18), AP13–Kanlow (702 cM vs. 638 cM; P = 0.13), and VS16–Kanlow (632 cM vs. 607 cM; P = 0.55) comparisons. The Okada Alamo (male) LGs were, however, significantly longer at the interval level compared to our VS16 (male) LGs (584 cM vs. 494 cM (P = 0.013).
Discussion
Linkage Maps
These are the first complete linkage maps generated in a full-sib population derived from a cross between a heterozygous lowland genotype (AP13, a clonally maintained plant selected from cultivar Alamo) and a heterozygous upland genotype (VS16, a Summer clone). Because we used an F1 population, we assessed recombination within AP13 and VS16 and our two maps therefore represent the lowland and upland switchgrass ecotype genomes. Within the lowland AP13 clone, we obtained 18 LGs that could be organized into nine homoeologous groups presumably corresponding to the nine homoeologous (A and B subgenome) switchgrass chromosomes. Grouping of the VS16 markers, however, only yielded 17 LGs. One-to-one homology was established between 15 VS16 LGs and 15 AP13 LGs. The two remaining VS16 LGs had no loci in common with any of the three remaining AP13 LGs, II-a, VII-a, and VIII-a. However, one had one marker (SWW2455) in common with VS16 LG II-b, and the other had two markers (NFSG102 and SWW2308) in common with VS16 LG VIII-b. The most likely explanation is that the two unassigned VS16 LGs are II-a and VIII-a, and that the common markers were identified by primer sets that amplified homoeologous regions in the A and the B genome that were polymorphic only in VS16.
In addition to those three markers, we identified a further 12 sets of loci that mapped to two homoeologous chromosomes, seven of which mapped to the A genome in AP13 and the B genome in VS16 or vice versa, four that mapped to A and B genomes in VS16, and one that mapped to the A and B genomes in AP13, bringing the number of loci sets that mapped to only homoeologous chromosomes to 13 %. We used the term ‘loci set’ for loci that were detected by the same primer pair, including single copy loci that mapped to homologous positions in AP13 and VS16 (homologous loci), single copy loci that mapped to homoeologous positions in the two switchgrass subgenomes (homoeologous loci) and duplicated loci that mapped to unrelated switchgrass chromosomes.
Excluding markers that mapped to unrelated chromosomes, 81 % of loci sets mapped to homologous chromosomes in the AP13 × VS16 mapping population, 14 % mapped to homoeologous chromosomes and 5 % mapped to both homologous and homoeologous chromosomes (ESM 4). If we also take the loci into account that were mapped in common between our maps and those generated by Okada and colleagues [11], the percentage of primers identifying loci that mapped only to homologous loci, only to homoeologous loci, and to both homologous and homoeologous loci is 83.5, 5.5, and 11 %, respectively (ESM 4). The fact that homoeologous loci (mapped on both A and B subgenomes) were mapped at a fivefold lower frequency than homologous loci (mapped in both AP13 and VS16) indicates that many of the primer sets are genome specific. This agrees with observations made in previous mapping studies [11, 14] and supports the contention that switchgrass is an allotetraploid.
Potential explanations for our inability to identify a VII-a LG in the upland genotype VS16 include (1) non-random distribution of the mapped markers; (2) mapping error (for example markers belonging to LG VII-a have been incorporated into another LG); (3) VS16 is an aneuploid and lacks the VII-a chromosome; and (4) LG VII-a is maintained largely in homozygous condition in VS16, leading to a lack of polymorphism for markers on this LG. The fact that the developed SSR markers are relatively evenly distributed over the 18 chromosomes in the AP13 map argues against non-random distribution of mapped markers as the cause of the absence of LG VII-a in the VS16 map. Incorporation of VII-a into another LG is also unlikely as none of the VS16 LGs, except VII-b-m, carry loci that are homologous or homoeologous to loci on VII-a-f, VII-b-f, or VII-b-m.
Aneuploidy has been observed in tetraploid as well as octoploid switchgrass, but the frequency is much higher in the latter [27]. The same authors also demonstrated through in situ hybridization of a ribosomal probe to mitotic metaphase chromosomes that many octoploids showed chromosomal rearrangements. No such rearrangements were observed in the tetraploids examined, but the resolution of this study was low since only a single probe was used [27]. Combining genomic and fluorescent in situ hybridization in a recently formed allotetraploid, Tragopogon miscellus, Chester and colleagues [28] showed that both rearrangements involving chromosomes from different subgenomes and compensating aneuploidy occurred at a high frequency in natural populations of this species. In a compensating allopolyploid, a chromosome from one of the subgenomes is replaced by its homoeologs so that the overall chromosome number remains unchanged. The DNA content of VS16 has been determined to be within the range expected for tetraploid switchgrass accessions [9] but, since no GISH or FISH have been performed on VS16, it is unknown whether this clone carries intersubgenome translocations. However, rearrangements and compensating aneuploidy in VS16 would likely lead to multivalent formation during meiosis resulting in tetrasomic inheritance of some markers. We therefore examined the segregation ratios in LG VII-b. LG VII-b consists of 12 markers, seven that segregate in a 1:1 ratio at the 5 % significance level, and five that deviated from a 1:1 ratio (P < 0.001). The segregation ratio of one of the distorted markers fits a 5:1 ratio, which would be expected of segregation of a double-dose dominant marker in an autotetraploid. The remainder of the distorted markers on VII-b, however, fits a 2:1 ratio to 3:1 ratio. Furthermore, the ratio of A:B alleles gradually increased from 1.12 to 6.23 from the top to the bottom of LG VII-b, which suggests that the distortion is caused by the presence of a ‘distortion factor’ rather than by tetrasomic inheritance. Other LGs containing clusters of markers with distorted segregation ratios are I-a and III-b in VS16.
Switchgrass is an outbreeding species, and most chromosome regions are expected to be in heterozygous condition. Tracts of extended homozygosity which, on average, span 1 to 2 Mb but can be as long as 17.9 Mb have been observed in humans [29, 30]. These tracts are found mainly in regions of high linkage disequilibrium and low recombination, and must be present in sufficiently high frequency in the population to sometimes being inherited from both parents by chance. Liu et al. [14] attributed homozygosity in switchgrass accession NL94 LYE, the line that was selfed to generate an F2 mapping population, as the underlying cause to the fact that only four closely linked markers were mapped to LG VII-b. We examined the profiles in AP13, VS16, and their progeny of nine SSRs that had been mapped to LG VII-a in the female parent, AP13, and that gave amplification patterns in which we could account for all major fragments. Four SSRs amplified at least one fragment in VS16 that was monomorphic, and three of those also amplified fragments in VS16 that significantly deviated from a 1:1 segregation ratio. A further four SSRs amplified no major fragments in VS16 and one SSR amplified fragments in VS16 that cosegregated with AP13 fragments. The latter can occur if alleles are preferentially amplified. The amplification profiles of these nine SSRs suggest that homozygosity may play a role, but is unlikely the sole reason for our inability to construct linkage group VII-a in VS16. It may be that a coalescing of several factors such as homozygosity, segregation distortion, preferential amplification, and non-random marker distribution, each of which by itself would have had a limited effect, led to linkage group VII-a to be missed in VS16.
Intraspecific Comparative Analyses
Three types of comparative analyses were conducted. Comparing the linkage maps obtained in the lowland genotype AP13 with those of the upland genotype VS16 showed that, with one exception, marker orders were completely colinear and there were no significant differences in the recombination rates between the two ecotypes. The significantly lower number of markers mapped per LG in VS16 compared to AP13 (0.001 ≤ P < 0.01) suggest that the overall level of heterozygosity is lower in VS16 than in AP13. Much of that difference is due to the lack of LG VII-a and the limited number of markers on II-a and VIII-a in the upland genotype VS16. However, even when omitting those three LGs, the number of loci mapped per LG is lower in VS16 than in AP13 (0.01 ≤ P < 0.05). Missaoui et al. [31] found in an analysis of 85 RFLP fragments in 14 Alamo genotypes including AP13 and three Summer genotypes including VS16 that the polymorphism level, measured as the percentage of fragments that were polymorphic between at least two genotypes, was higher in Summer (64 %) than in Alamo (60 %). This measure might have underestimated the overall variation in Summer compared to Alamo since it does not take into account the larger number of Alamo genotypes analyzed. On the other hand, a similar study using 55 SSR markers and 16 genotypes each of Alamo and Summer, identified 199 alleles including 19 private alleles in Alamo and 181 (one private) alleles in Summer, indicating a lower level of variation in Summer populations compared to Alamo populations [32]. Since the number of markers per LG varies over a much wider range in VS16 (sixfold range; excluding the missing LG) compared to AP13 (twofold range), it would be interesting to specifically look at haplotype diversity at a chromosomal level in Summer populations.
The incorporation of markers previously mapped by Okada et al. [11] also allowed us to compare our AP13 and VS16 maps with the Okada Alamo and Kanlow maps. Notwithstanding a few differences that were most easily explained by scoring errors (e.g., inversion of two closely linked markers) or paralogy, complete colinearity was observed. Recombination in intervals defined by common loci was not significantly different except between VS16 and Alamo. However, there might not be a biological reason for this difference as Okada et al. [11] had noted that the longer map length of Alamo could be an artifact due to biased recombination estimates caused by interactions between markers displaying distorted segregation ratios. As in the Okada maps, regions of severely distorted marker ratios were identified in the male maps (Fig. 1). Of the three male LGs showing transmission distortion in our maps (I-a, III-b, and VII-b), two also showed segregation distortion in the Okada maps. While regions of segregation distortion are often cross-dependent, it is possible that chromosomes I-a and VII-b carry loci that affect allelic transmission. Okada et al. [11] hypothesized that regions of segregation distortion might carry orthologs to the self-incompatibility loci Z and S. Using comparative information, they found that the distorted region on LG VII-b, which is also distorted in our maps, is orthologous to the region in rye carrying the Z locus. Similarly, the S locus was tentatively placed on homology group III. Although this homology group did not carry any regions showing distorted segregation ratios in the Okada maps, chromosome III-b is one of the three male LGs with distorted ratio in our maps. The presence of self-incompatibility loci would lead to gametes contributed through the pollen and carrying an incompatible allele being excluded, and hence would also explain why the distortion is observed in the male and not the female maps.
Conclusions
The first complete genetic maps generated in a cross between a lowland and an upland switchgrass ecotype provide a valuable tool for the identification and manipulation in breeding programs of QTL that differentiate the two ecotypes such as stem thickness, spring regrowth, biomass yield, and cold tolerance, assuming that those traits are not fixed within ecotypes. The maps show that marker orders, levels of recombination, and distribution of recombination events are highly similar between the upland and lowland ecotypes. This suggests that chromosomal exchanges between the two ecotypes should occur freely and hence introgression of favorable traits from the higher biomass yielding lowlands into the more cold-tolerant uplands and vice versa should be an attainable breeding goal. Furthermore, the mapped markers will provide important anchor points for assembling the switchgrass genome sequence. Because switchgrass is an outcrossing tetraploid, up to four haplotypes may be obtained at every locus, which greatly complicates sequence assembly. The genetic maps will assist with organizing the sequence contigs by LG and by subgenome, and are one more tool in the quest for a fully assembled switchgrass genome.
References
Sanderson MA, Reed RL, McLaughlin SB, Wullschleger SD, Conger BV et al (1996) Switchgrass as a sustainable bioenergy crop. Bioresource Technol 56:83–93
McLaughlin SB, Kszos LA (2005) Development of switchgrass (Panicum virgatum) as a bioenergy feedstock in the United States. Biomass Bioenergy 28:515–535
Alexopoulou E, Sharma N, Papatheohari Y, Christou M, Piscioneri I, Panoutsou C, Pignatelli V (2008) Biomass yields for upland and lowland switchgrass varieties grown in the Mediterranean region. Biomass Bioenergy 32:926–933
Christian DC, Elberson HW (1998) Energy plant species: their use and impact on environment and development. In: Bassam NE (ed) Switchgrass. James, London, pp 257–263
Moser LE, Vogel KP (1995) Switchgrass, big bluestem and indiangrass. In: Barnes RF (ed) Forages, vol 1, An introduction to grassland agriculture. Iowa State University Press, Ames, pp 409–420
Jr Porter CL (1966) An analysis of variation between upland and lowland switchgrass, Panicum virgatum L., in central Oklahoma. Ecology 47:980–992
Gunter LE, Tuskan GA, Wullschleger SD (1996) Diversity among populations of switchgrass based on RAPD markers. Crop Sci 36:1017–1022
Hopkins AA, Taliaferro CM, Murphy CD, Christian D (1996) Chromosome number and nuclear DNA content of several switchgrass populations. Crop Sci 36:1192–1195
Narasimhamoorthy B, Saha MC, Swaller T, Bouton JH (2008) Genetic diversity in switchgrass collections assessed by EST-SSR markers. Bioenergy Research 1:136–146
Missaoui AM, Paterson AH, Bouton JH (2005) Investigation of genomic organization in switchgrass (Panicum virgatum L.) using DNA markers. Theor Appl Genet 110:1372–1383
Okada M, Lanzatella C, Saha MC, Bouton J, Wu R, Tobias CM (2010) Complete switchgrass genetic maps reveal subgenome collinearity, preferential pairing and multilocus interactions. Genetics 185:745–760
Triplett JK, Wang Y, Zhong J, Kellogg EA (2012) Five nuclear loci resolve the polyploid history of switchgrass (Panicum virgatum L.) and relatives. PLoS One 7:e38702
Vogel KP, Jung HJG (2001) Genetic modification of herbaceous plants for feed and fuel. Crit Rev Plant Sci 20:15–49
Liu L, Wu Y, Wang Y, Samuels T (2012) A high-density simple sequence repeat-based genetic linkage map of switchgrass. G3 2:357–370
Zane L, Bargelloni L, Patarnello T (2002) Strategies for microsatellite isolation: a review. Mol Ecol 11:1–16
Dida MM, Srinivasachary RS, Bennetzen JL, Gale MD, Devos KM (2007) The genetic map of finger millet, Eleusine coracana. Theor Appl Genet 114:321–332
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana, Totowa, pp 365–386
Tobias CM, Hayden DM, Twigg P, Sarath G (2006) Genic microsatellite markers derived from EST sequences of switchgrass (Panicum virgatum L.). Mol Ecol Notes 6:185–187
Tobias CM, Sarath G, Twigg P, Lindquist E, Pangilinan J et al (2008) Comparative genomics in switchgrass using 61,585 high-quality expressed sequence tags. The Plant Genome 1:111–124
Jaccoud D, Peng K, Feinstein D, Kilian A (2001) Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res 29:e25–e31
Kilian A, Wenzl P, Huttner E, Carling J, Xia L et al (2012) Diversity arrays technology: a generic genome profiling technology on open platforms. Methods Mol Biol 888:67–89
Van Ooijen JW (2006) Joinmap® 4, Software for the calculation of genetic linkage maps in experimental populations
Grattapaglia D, Sederoff R (1994) Genetic linkage maps of Eucalyptus grandis and Eucalyptus uropylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics 137:1121–1137
Kosambi DD (1944) The estimation of map values from recombination value. Ann Eugen 12:172–175
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Lincoln S, Daly M, Lander ES (1993) Constructing genetic maps with MAPMAKER/EXP 3.0. Whitehead Institute for Biomedical Research, Cambridge, Massachusetts, USA
Costich D, Friebe B, Sheehan MJ, Casler MD, Buckler ES (2010) Genome-size variation in switchgrass (Panicum virgatum): flow cytometry and cytology reveal rampant aneuploidy. Plant Gen 3:130–141
Chester M, Gallagher JP, Symonds VV, Cruz da Silva AV, Mavrodiev EV, Leitch AR, Soltis PS, Soltis DE (2012) Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proc Natl Acad Sci 109:76–1181
Gibson J, Morton NE, Collins A (2006) Extended tracts of homozygosity in outbred human populations. Hum Mol Genet 15:789–795
Nalls MA, Simon-Sanchez J, Gibbs JR, Paisan-Ruiz C, Bras JT et al (2009) Measures of autozygosity in decline: globalization, urbanization, and its implications for medical genetics. PLoS Genet 5:e1000415
Missaoui AM, Paterson AH, Bouton JH (2006) Molecular markers for the classification of switchgrass (Panicum virgatum L.) germplasm and to assess genetic diversity in three synthetic switchgrass populations. Genet Resour Crop Evol 53:1291–1302
Zalapa JE, Price DL, Kaeppler SM, Tobias CM, Okada M, Casler MD (2011) Hierarchical classification of switchgrass genotypes using SSR and chloroplast sequences: ecotypes, ploidies, gene pools, and cultivars. Theor Appl Genet 122:805–817
Acknowledgments
Funding for this research was provided by the Office of Biological and Environmental Research in the Department of Energy (DOE) Office of Science to the BioEnergy Science Center (BESC) and by the National Institute of Food And Agriculture Plant Feedstock Genomics for Bioenergy Program (grant no. 2007-35504-18246).
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Desalegn Serba, Limin Wu, and Guillaume Daverdin contributed equally to the work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(XLS 92 kb)
ESM 2
Comparison of the genetic maps generated with the software programs Mapmaker (left hand maps) and JoinMap (right hand maps). For marker orders that are different in the Mapmaker and JoinMap maps, the LOD score for the order observed for a particular group of markers, indicated with a vertical bar, in the JoinMap maps compared to that in the Mapmaker maps is given on the right-handside of the JoinMap maps. If the JoinMap order was not one of the 40 most likely orders as determined by the mapmaker ‘compare’ command, the LOD score for the JoinMap marker order was given as less than the LOD score calculated for the 40th marker order (PDF 1.81 MB)
ESM 3
(XLS 2559 kb)
ESM 4
(XLS 112 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Serba, D., Wu, L., Daverdin, G. et al. Linkage Maps of Lowland and Upland Tetraploid Switchgrass Ecotypes. Bioenerg. Res. 6, 953–965 (2013). https://doi.org/10.1007/s12155-013-9315-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12155-013-9315-6