Abstract
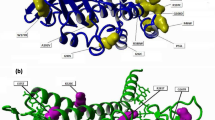
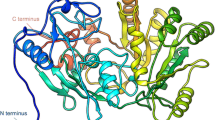
The enzyme 3-hydroxy-3-methyl-glutaryl-CoA reductase (HMG-CoA-R) is the fundamental target for the treatment of hypercholesterolemia nowadays. The HMG-CoA-R clinical active site inhibitors (statins) are among the most widespread and profitable drugs ever sold but their side effects (myopathies, sometimes severe) still limit their use, which makes the finding of alternatives to statins a field of intense research. In this line, we address here a new strategy for inhibiting the homotetrameric HMG-CoA-R. The enzyme consists of a "dimer of dimers”, each dimer having two active sites. We pursue here the inhibition of enzyme oligomerization, through drug binding to the dimer interface. We have computationally mutated 232 interfacial residues by alanine and calculated the loss in binding free energy among the monomers that build up each dimer of the homotetramer. This led to the identification of the (ten) key residues for the formation of the active dimer (Glu528, Ile531, Met534, Tyr644, Glu665, Asn686, Lys692, Lys735, Met742, and Val863). The results show that these residues are located in two specific spots of the protein with a cleft shape, whose shape and size is favorable for small drug binding. It is expectable that small molecules specifically bound to these druggable pockets will have a major effect on the oligomerization of the protein or/and in active site formation. This paves the way for the discovery of new families of inhibitors of HMG-CoA-R.







Similar content being viewed by others
References
Black DM (2003) Therapeutic targets in cardiovascular disease: a case for high-density lipoprotein cholesterol. Am J Cardiol 91:40E–43E
Parks LW (1978) Metabolism of Sterols in Yeast. CRC Crit Rev Microbiol 6:301–341
Goldstein JL, Brown MS (1990) Regulation of the mevalonate pathway. Nature 343:425–430
Chappell J, Wolf F, Proulx J, Cuellar R, Saunders C (1995) Is the reaction catalyzed by 3-hydroxy-3-methylglutaryl coenzyme-a reductase a rate-limiting step for isoprenoid biosynthesis in plants. Plant Physiol 109:1337–1343
Istvan ES, Deisenhofer J (2001) Structural mechanism for statin inhibition of HMG-CoA reductase. Science 292:1160–1164
Brown MS, Faust JR, Goldstein JL, Kaneko I, Endo A (1978) Induction of 3-hydroxy-3-methylglutaryl coenzyme-a reductase-activity in human fibroblasts incubated with compactin (Ml-236b), a competitive inhibitor of reductase. J Biol Chem 253:1121–1128
Singh N, Tamariz J, Chamorro G, Medina-Franco JL (2009) Inhibitors of HMG-CoA reductase: current and future prospects. Mini-Rev Med Chem 9:1272–1283
Istvan E (2003) Statin inhibition of HMG-CoA reductase: a 3-dimensional view. Atheroscler Suppl 4:3–8
Hermann M, Bogsrud MP, Molden E, Asberg A, Mohebi BU, Ose L, Retterstol K (2006) Exposure of atorvastatin is unchanged but lactone and acid metabolites are increased several-fold in patients with atorvastatin-induced myopathy. Clin Pharmacol Ther 79:532–539
Golomb BA, Evans MA (2008) Statin adverse effects: a review of the literature and evidence for a mitochondrial mechanism. Am J Cardiovasc Drugs 8:373–418
Skottheim IB, Gedde-Dahl A, Hejazifar S, Hoel K, Asbery A (2008) Statin induced myotoxicity: the lactone forms are more potent than the acid forms in human skeletal muscle cells in vitro. Eur J Pharm Sci 33:317–325
Furberg CD, Pitt B (2001) Withdrawal of cerivastatin from the world market. Curr Control Trials Cardiovasc Med 2:205–207
Graham DJ, Staffa JA, Shatin D, Andrade SE, Schech SD, La Grenade L, Gurwitz JH, Chan KA, Goodman MJ, Platt R (2004) Incidence of hospitalized rhabdomyolysis in patients treated with lipid-lowering drugs. JAMA-J Am Med Assoc 292:2585–2590
Keskin O, Gursoy A, Ma B, Nussinov R (2008) Principles of protein-protein interactions: what are the preferred ways for proteins to interact? Chem Rev 108:1225–1244
Arkin MR, Wells JA (2004) Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov 3:301–317
Chene P (2006) Drugs targeting protein-protein interactions. Chem Med Chem 1:400–411
Wells JA, McClendon CL (2007) Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature 450:1001–1009
Zinzalla G, Thurston DE (2009) Targeting protein-protein interactions for therapeutic intervention: a challenge for the future. Futur Med Chem 1:65–93
Pieraccini S, Saladino G, Cappelletti G, Cartelli D, Francescato P, Speranza G, Manitto P, Sironi M (2009) In silico design of tubulin-targeted antimitotic peptides. Nat Chem 1:642–648
Massova I, Kollman PA (1999) Computational alanine scanning to probe protein − protein interactions: a novel approach to evaluate binding free energies. J Am Chem Soc 121:8133–8143
Kortemme T, Baker D (2002) A simple physical model for binding energy hot spots in protein-protein complexes. Proc Natl Acad Sci U S A 99:14116–14121
Istvan ES, Deisenhofer J (2000) The structure of the catalytic portion of human HMG-CoA reductase. Biochim Biophys Acta 1529:9–18
Ribeiro JV, Cerqueira NMFSA, Moreira IS, Fernandes PA, Ramos MJ (2012) CompASM: an Amber-VMD alanine scanning mutagenesis plug-in. Theor Chem Acc 131:1271
Moreira IS, Fernandes PA, Ramos MJ (2007) Computational alanine scanning mutagenesis - an improved methodological approach. J Comput Chem 28:644–654
Moreira IS, Fernandes PA, Ramos MJ (2007) Hot spots-a review of the protein-protein interface determinant amino-acid residues. Protein-Struct Funct Bioinforma 68:803–812
Moreira IS, Fernandes PA, Ramos MJ (2007) Hot spot occlusion from bulk water: a comprehensive study of the complex between the lysozyme HEL and the antibody FVD1.3. J Phys Chem B 111:2697–2706
Sousa SF, Tamames B, Fernandes PA, Ramos MJ (2011) Detailed atomistic analysis of the HIV-1 protease interface. J Phys Chem B 115:7045–7057
Istvan ES, Deisenhofer J (2000) The structure of the catalytic portion of human HMG-CoA reductase. Biochim Biophys Acta Mol Cell Biol Lipids 1529:9–18
Li H, Robertson AD, Jensen JH (2005) Very fast empirical prediction and rationalization of protein pK(a) values. Protein Struct Funct Bioinforma 61:704–721
Olsson MHM, Sondergaard CR, Rostkowski M, Jensen JH (2011) PROPKA3: consistent treatment of internal and surface residues in empirical pK(a) predictions. J Chem Theory Comput 7:525–537
Case DA, Darden TA, Cheatham, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA (2006) AMBER9, San Francisco
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) Numerical-integration of Cartesian equations of motion of a system with constraints-molecular-dynamics of N-alkanes. J Comput Phys 23:327–341
Martins SA, Perez MAS, Moreira IS, Sousa SF, Ramos MJ, Fernandes PA (2013) Computational alanine scanning mutagenesis: MM-PBSA vs TI. J Chem Theory Comput 9:1311–1319
Moreira IS, Fernandes PA, Ramos MJ (2006) Unraveling the importance of protein-protein interaction: application of a computational alanine-scanning mutagenesis to the study of the IgG1 streptococcal protein G (C2 fragment) complex. J Phys Chem B 110:10962–10969
Moreira IS, Fernandes PA, Ramos MJ (2008) Protein-protein recognition: a computational mutagenesis study of the MDM2-P53 complex. Theor Chem Accounts 120:533–542
Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE 3rd (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 33:889–897
Huo S, Massova I, Kollman PA (2002) Computational alanine scanning of the 1:1 human growth hormone-receptor complex. J Comput Chem 23:15–27
Humphrey W, Dalke A, Schulten K (1996) VMD-visual molecular dynamics. J Mol Graph 14:33–38
Ribeiro JV, Cerqueira NMFSA, Fernandes PA, Ramos MJ (2013) VOLAREA - a Bioinformatic tool to calculate the surface area and the volume of molecular systems. Chem Biol 82:743–755 doi: 10.1111/cbdd.12197
Acknowledgments
Diana Gesto is grateful to the Fundacao para a Ciencia e Technologia (FCT), Portugal for a PhD Grant (SFRH/BPD/44469/2008 - Mudar) and N.M.F.S.A. Cerqueira to program Ciência 2007. This work has been funded by FEDER/COMPETE and Fundação para a Ciência e a Tecnologia through grant n.º PTDC/QUI-QUI/102760/2008and grant no. PEst-C/EQB/LA0006/2011
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gesto, D.S., Cerqueira, N.M.F.S.A., Ramos, M.J. et al. Discovery of new druggable sites in the anti-cholesterol target HMG-CoA reductase by computational alanine scanning mutagenesis. J Mol Model 20, 2178 (2014). https://doi.org/10.1007/s00894-014-2178-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-014-2178-8