Abstract
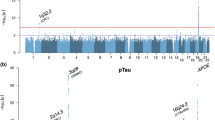
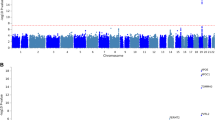
More than 20 genetic loci have been associated with risk for Alzheimer’s disease (AD), but reported genome-wide significant loci do not account for all the estimated heritability and provide little information about underlying biological mechanisms. Genetic studies using intermediate quantitative traits such as biomarkers, or endophenotypes, benefit from increased statistical power to identify variants that may not pass the stringent multiple test correction in case–control studies. Endophenotypes also contain additional information helpful for identifying variants and genes associated with other aspects of disease, such as rate of progression or onset, and provide context to interpret the results from genome-wide association studies (GWAS). We conducted GWAS of amyloid beta (Aβ42), tau, and phosphorylated tau (ptau181) levels in cerebrospinal fluid (CSF) from 3146 participants across nine studies to identify novel variants associated with AD. Five genome-wide significant loci (two novel) were associated with ptau181, including loci that have also been associated with AD risk or brain-related phenotypes. Two novel loci associated with Aβ42 near GLIS1 on 1p32.3 (β = −0.059, P = 2.08 × 10−8) and within SERPINB1 on 6p25 (β = −0.025, P = 1.72 × 10−8) were also associated with AD risk (GLIS1: OR = 1.105, P = 3.43 × 10−2), disease progression (GLIS1: β = 0.277, P = 1.92 × 10−2), and age at onset (SERPINB1: β = 0.043, P = 4.62 × 10−3). Bioinformatics indicate that the intronic SERPINB1 variant (rs316341) affects expression of SERPINB1 in various tissues, including the hippocampus, suggesting that SERPINB1 influences AD through an Aβ-associated mechanism. Analyses of known AD risk loci suggest CLU and FERMT2 may influence CSF Aβ42 (P = 0.001 and P = 0.009, respectively) and the INPP5D locus may affect ptau181 levels (P = 0.009); larger studies are necessary to verify these results. Together the findings from this study can be used to inform future AD studies.


Similar content being viewed by others
References
Adams HH, Hibar DP, Chouraki V, Stein JL, Nyquist PA, Renteria ME, Trompet S, Arias-Vasquez A, Seshadri S, Desrivieres S et al (2016) Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat Neurosci. doi:10.1038/nn.4398
Andersson C, Blennow K, Almkvist O, Andreasen N, Engfeldt P, Johansson SE, Lindau M, Eriksdotter-Jonhagen M (2008) Increasing CSF phospho-tau levels during cognitive decline and progression to dementia. Neurobiol Aging 29:1466–1473. doi:10.1016/j.neurobiolaging.2007.03.027
Anttila V, Bulik-Sullivan B, Finucane HK, Bras J, Duncan L, Escott-Price V, Falcone G, Gormley P, Malik R, Patsopoulos N, et al (2016) Analysis of shared heritability in common disorders of the brain. bioRxiv doi:10.1101/048991
Baik SH, Cha MY, Hyun YM, Cho H, Hamza B, Kim DK, Han SH, Choi H, Kim KH, Moon M et al (2014) Migration of neutrophils targeting amyloid plaques in Alzheimer’s disease mouse model. Neurobiol Aging 35:1286–1292. doi:10.1016/j.neurobiolaging.2014.01.003
Balestrini A, Cosentino C, Errico A, Garner E, Costanzo V (2010) GEMC1 is a TopBP1-interacting protein required for chromosomal DNA replication. Nat Cell Biol 12:484–491. doi:10.1038/ncb2050
Benitez BA, Cooper B, Pastor P, Jin SC, Lorenzo E, Cervantes S, Cruchaga C (2013) TREM2 is associated with the risk of Alzheimer’s disease in Spanish population. Neurobiol Aging 34(1711):e1715–e1717. doi:10.1016/j.neurobiolaging.2012.12.018
Blennow K, Hampel H, Weiner M, Zetterberg H (2010) Cerebrospinal fluid and plasma biomarkers in Alzheimer disease. Nat Rev Neurol 6:131–144. doi:10.1038/nrneurol.2010.4
Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S et al (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 22:1790–1797. doi:10.1101/gr.137323.112
Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, Duncan L, ReproGen C, Psychiatric Genomics C, Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control C et al (2015) An atlas of genetic correlations across human diseases and traits. Nat Genet 47:1236–1241. doi:10.1038/ng.3406
Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, Daly MJ, Price AL, Neale BM, Schizophrenia Working Group of the Psychiatric Genomics C (2015) LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47:291–295. doi:10.1038/ng.3211
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:7. doi:10.1186/s13742-015-0047-8
Cross-Disorder Group of the Psychiatric Genomics C (2013) Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 45:984–994. doi:10.1038/ng.2711
Cruchaga C, Kauwe JS, Harari O, Jin SC, Cai Y, Karch CM, Benitez BA, Jeng AT, Skorupa T, Carrell D et al (2013) GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer’s disease. Neuron 78:256–268. doi:10.1016/j.neuron.2013.02.026
Cruchaga C, Kauwe JS, Mayo K, Spiegel N, Bertelsen S, Nowotny P, Shah AR, Abraham R, Hollingworth P, Harold D et al (2010) SNPs associated with cerebrospinal fluid phospho-tau levels influence rate of decline in Alzheimer’s disease. PLoS Genet 6:e1001101. doi:10.1371/journal.pgen.1001101
Cruchaga C, Kauwe JS, Nowotny P, Bales K, Pickering EH, Mayo K, Bertelsen S, Hinrichs A, Alzheimer’s Disease Neuroimaging I, Fagan AM et al (2012) Cerebrospinal fluid APOE levels: an endophenotype for genetic studies for Alzheimer’s disease. Hum Mol Genet 21:4558–4571. doi:10.1093/hmg/dds296
de Leon MJ, DeSanti S, Zinkowski R, Mehta PD, Pratico D, Segal S, Clark C, Kerkman D, DeBernardis J, Li J et al (2004) MRI and CSF studies in the early diagnosis of Alzheimer’s disease. J Intern Med 256:205–223. doi:10.1111/j.1365-2796.2004.01381.x
De Meyer G, Shapiro F, Vanderstichele H, Vanmechelen E, Engelborghs S, De Deyn PP, Coart E, Hansson O, Minthon L, Zetterberg H et al (2010) Diagnosis-independent Alzheimer disease biomarker signature in cognitively normal elderly people. Arch Neurol 67:949–956. doi:10.1001/archneurol.2010.179
Delaneau O, Marchini J, Consortium GP (2014) Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat Commun. doi:10.1038/ncomms4934
Deming Y, Xia J, Cai Y, Lord J, Del-Aguila JL, Fernandez MV, Carrell D, Black K, Budde J, Ma S et al (2016) Genetic studies of plasma analytes identify novel potential biomarkers for several complex traits. Scientific Reports 6:18092. doi:10.1038/srep18092
Deming Y, Xia J, Cai Y, Lord J, Holmans P, Bertelsen S, Holtzman D, Morris JC, Bales K, Pickering EH et al (2016) A potential endophenotype for Alzheimer’s disease: cerebrospinal fluid clusterin. Neurobiol Aging 37(208):e201–e209. doi:10.1016/j.neurobiolaging.2015.09.009
Dudbridge F (2016) Polygenic epidemiology. Genet Epidemiol 40:268–272. doi:10.1002/gepi.21966
Escott-Price V, Shoai M, Pither R, Williams J, Hardy J (2017) Polygenic score prediction captures nearly all common genetic risk for Alzheimer’s disease. Neurobiol Aging 49:214-e217–214-e211. doi:10.1016/j.neurobiolaging.2016.07.018
Escott-Price V, Sims R, Bannister C, Harold D, Vronskaya M, Majounie E, Badarinarayan N, Gerad Perades, Morgan K, Gerad/Perades, consortia I, Consortia I et al (2015) Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain 138:3673–3684. doi:10.1093/brain/awv268
Euesden J, Lewis CM, O’Reilly PF (2015) PRSice: polygenic Risk Score software. Bioinformatics 31:1466–1468. doi:10.1093/bioinformatics/btu848
Fagan AM, Head D, Shah AR, Marcus D, Mintun M, Morris JC, Holtzman DM (2009) Decreased cerebrospinal fluid Aβ(42) correlates with brain atrophy in cognitively normal elderly. Ann Neurol 65:176–183. doi:10.1002/ana.21559
Fagan AM, Roe CM, Xiong C, Mintun MA, Morris JC, Holtzman DM (2007) Cerebrospinal fluid tau/β-amyloid(42) ratio as a prediction of cognitive decline in nondemented older adults. Arch Neurol 64:343–349. doi:10.1001/archneur.64.3.noc60123
Farfel JM, Yu L, De Jager PL, Schneider JA, Bennett DA (2016) Association of APOE with tau-tangle pathology with and without β-amyloid. Neurobiol Aging 37:19–25. doi:10.1016/j.neurobiolaging.2015.09.011
Farley K, Stolley JM, Zhao P, Cooley J, Remold-O’Donnell E (2012) A serpinB1 regulatory mechanism is essential for restricting neutrophil extracellular trap generation. J Immunol 189:4574–4581. doi:10.4049/jimmunol.1201167
Farrer LA, Cupples L, Haines JL et al (1997) Effects of age, sex, and ethnicity on the association between apolipoprotein e genotype and alzheimer disease: a meta-analysis. JAMA 278:1349–1356. doi:10.1001/jama.1997.03550160069041
Fleming LM, Weisgraber KH, Strittmatter WJ, Troncoso JC, Johnson GV (1996) Differential binding of apolipoprotein E isoforms to tau and other cytoskeletal proteins. Exp Neurol 138:252–260. doi:10.1006/exnr.1996.0064
Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, Fiske A, Pedersen NL (2006) Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiat 63:168–174. doi:10.1001/archpsyc.63.2.168
Grundberg E, Adoue V, Kwan T, Ge B, Duan QL, Lam KCL, Koka V, Kindmark A, Weiss ST, Tantisira K et al (2011) Global analysis of the impact of environmental perturbation on cis-regulation of gene expression. PLoS Genet. doi:10.1371/journal.pgen.1001279
GTEx Consortium (2015) Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348:648–660. doi:10.1126/science.1262110
Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JSK, Lupton MK et al (2013) TREM2 variants in Alzheimer’s disease. New Engl J Med 368:117–127. doi:10.1056/NEJMoa1211851
Guo H, Fortune MD, Burren OS, Schofield E, Todd JA, Wallace C (2015) Integration of disease association and eQTL data using a Bayesian colocalisation approach highlights six candidate causal genes in immune-mediated diseases. Hum Mol Genet 24:3305–3313. doi:10.1093/hmg/ddv077
Han MR, Schellenberg GD, Wang LS, Alzheimer’s Disease Neuroimaging I (2010) Genome-wide association reveals genetic effects on human Aβ42 and tau protein levels in cerebrospinal fluids: a case control study. BMC Neurol 10:90. doi:10.1186/1471-2377-10-90
Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A et al (2009) Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease (vol 41, pg 1088, 2009). Nat Genet 41:1156. doi:10.1038/ng1009-1156d
Heppner FL, Ransohoff RM, Becher B (2015) Immune attack: the role of inflammation in Alzheimer disease. Nat Rev Neurosci 16:358–372. doi:10.1038/nrn3880
Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V et al (2011) Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet 43:429–435. doi:10.1038/ng.803
Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR (2012) Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 44:955. doi:10.1038/ng.2354
Hulette CM, Welsh-Bohmer KA, Murray MG, Saunders AM, Mash DC, McIntyre LM (1998) Neuropathological and neuropsychological changes in “normal” aging: evidence for preclinical Alzheimer disease in cognitively normal individuals. J Neuropathol Exp Neurol 57:1168–1174
Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ et al (2013) Variant of TREM2 associated with the risk of Alzheimer’s disease. New Engl J Med 368:107–116. doi:10.1056/NEJMoa1211103
Kanai M, Matsubara E, Isoe K, Urakami K, Nakashima K, Arai H, Sasaki H, Abe K, Iwatsubo T, Kosaka T et al (1998) Longitudinal study of cerebrospinal fluid levels of tau, A beta1-40, and A beta1-42(43) in Alzheimer’s disease: a study in Japan. Ann Neurol 44:17–26. doi:10.1002/ana.410440108
Kauwe JS, Cruchaga C, Bertelsen S, Mayo K, Latu W, Nowotny P, Hinrichs AL, Fagan AM, Holtzman DM, Alzheimer’s Disease Neuroimaging I et al (2010) Validating predicted biological effects of Alzheimer’s disease associated SNPs using CSF biomarker levels. J Alzheimers Dis 21:833–842. doi:10.3233/JAD-2010-091711
Kauwe JS, Cruchaga C, Mayo K, Fenoglio C, Bertelsen S, Nowotny P, Galimberti D, Scarpini E, Morris JC, Fagan AM et al (2008) Variation in MAPT is associated with cerebrospinal fluid tau levels in the presence of amyloid-beta deposition. Proc Natl Acad Sci USA 105:8050–8054. doi:10.1073/pnas.0801227105
Kim S, Swaminathan S, Shen L, Risacher SL, Nho K, Foroud T, Shaw LM, Trojanowski JQ, Potkin SG, Huentelman MJ et al (2011) Genome-wide association study of CSF biomarkers Aβ1-42, t-tau, and p-tau181p in the ADNI cohort. Neurology 76:69–79. doi:10.1212/WNL.0b013e318204a397
Kyrousi C, Arbi M, Pilz GA, Pefani DE, Lalioti ME, Ninkovic J, Gotz M, Lygerou Z, Taraviras S (2015) Mcidas and GemC1 are key regulators for the generation of multiciliated ependymal cells in the adult neurogenic niche. Development 142:3661–3674. doi:10.1242/dev.126342
Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B et al (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45:1452–1458. doi:10.1038/ng.2802
Leoni V, Solomon A, Kivipelto M (2010) Links between ApoE, brain cholesterol metabolism, tau and amyloid β-peptide in patients with cognitive impairment. Biochem Soc Trans 38:1021–1025. doi:10.1042/BST0381021
Lepretre C, Tchakarska G, Blibech H, Lebon C, Torriglia A (2013) Apoptosis-inducing factor (AIF) and leukocyte elastase inhibitor/L-DNase II (LEI/LDNaseII), can interact to conduct caspase-independent cell death. Apoptosis 18:1048–1059. doi:10.1007/s10495-013-0862-2
Li QS, Parrado AR, Samtani MN, Narayan VA, Alzheimer’s Disease Neuroimaging I (2015) Variations in the FRA10AC1 Fragile Site and 15q21 Are Associated with Cerebrospinal Fluid Aβ1-42 Level. PLoS ONE 10:e0134000. doi:10.1371/journal.pone.0134000
Liraz O, Boehm-Cagan A, Michaelson DM (2013) ApoE4 induces Aβ42, tau, and neuronal pathology in the hippocampus of young targeted replacement apoE4 mice. Mol Neurodegener 8:16. doi:10.1186/1750-1326-8-16
Louwersheimer E, Wolfsgruber S, Espinosa A, Lacour A, Heilmann-Heimbach S, Alegret M, Hernandez I, Rosende-Roca M, Tarraga L, Boada M et al (2016) Alzheimer’s disease risk variants modulate endophenotypes in mild cognitive impairment. Alzheimers Dement 12:872–881. doi:10.1016/j.jalz.2016.01.006
Martiskainen H, Helisalmi S, Viswanathan J, Kurki M, Hall A, Herukka SK, Sarajarvi T, Natunen T, Kurkinen KM, Huovinen J et al (2015) Effects of Alzheimer’s disease-associated risk loci on cerebrospinal fluid biomarkers and disease progression: a polygenic risk score approach. J Alzheimers Dis 43:565–573. doi:10.3233/JAD-140777
Morris JC, Price JL (2001) Pathologic correlates of nondemented aging, mild cognitive impairment, and early-stage Alzheimer’s disease. J Mol Neurosci 17:101–118
Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK et al (2011) Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet 43:436–441. doi:10.1038/ng.801
O’Bryant SE, Lacritz LH, Hall J, Waring SC, Chan W, Khodr ZG, Massman PJ, Hobson V, Cullum CM (2010) Validation of the new interpretive guidelines for the clinical dementia rating scale sum of boxes score in the national Alzheimer’s coordinating center database. Arch Neurol 67:746–749. doi:10.1001/archneurol.2010.115
O’Bryant SE, Waring SC, Cullum CM, Hall J, Lacritz L, Massman PJ, Lupo PJ, Reisch JS, Doody R, Texas Alzheimer’s Research C (2008) Staging dementia using Clinical Dementia Rating Scale Sum of Boxes scores: a Texas Alzheimer’s research consortium study. Arch Neurol 65:1091–1095. doi:10.1001/archneur.65.8.1091
Olsson B, Lautner R, Andreasson U, Ohrfelt A, Portelius E, Bjerke M, Holtta M, Rosen C, Olsson C, Strobel G et al (2016) CSF and blood biomarkers for the diagnosis of Alzheimer’s disease: a systematic review and meta-analysis. Lancet Neurol 15:673–684. doi:10.1016/S1474-4422(16)00070-3
Piccio L, Deming Y, Del-Aguila JL, Ghezzi L, Holtzman DM, Fagan AM, Fenoglio C, Galimberti D, Borroni B, Cruchaga C (2016) Cerebrospinal fluid soluble TREM2 is higher in Alzheimer disease and associated with mutation status. Acta Neuropathol 131:925–933. doi:10.1007/s00401-016-1533-5
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909. doi:10.1038/ng1847
Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26:2336–2337. doi:10.1093/bioinformatics/btq419
Ramirez A, van der Flier WM, Herold C, Ramonet D, Heilmann S, Lewczuk P, Popp J, Lacour A, Drichel D, Louwersheimer E et al (2014) SUCLG2 identified as both a determinator of CSF Aβ1-42 levels and an attenuator of cognitive decline in Alzheimer’s disease. Hum Mol Genet 23:6644–6658. doi:10.1093/hmg/ddu372
Ridge PG, Hoyt KB, Boehme K, Mukherjee S, Crane PK, Haines JL, Mayeux R, Farrer LA, Pericak-Vance MA, Schellenberg GD et al (2016) Assessment of the genetic variance of late-onset Alzheimer’s disease. Neurobiol Aging 41(200):e213–e220. doi:10.1016/j.neurobiolaging.2016.02.024
Sabuncu MR, Buckner RL, Smoller JW, Lee PH, Fischl B, Sperling RA, Alzheimer’s Disease Neuroimaging I (2012) The association between a polygenic Alzheimer score and cortical thickness in clinically normal subjects. Cereb Cortex 22:2653–2661. doi:10.1093/cercor/bhr348
Sampson JN, Wheeler WA, Yeager M, Panagiotou O, Wang Z, Berndt SI, Lan Q, Abnet CC, Amundadottir LT, Figueroa JD et al (2015) Analysis of heritability and shared heritability based on genome-wide association studies for thirteen cancer types. J Natl Cancer Inst 107:djv279. doi:10.1093/jnci/djv279
Selkoe DJ, Hardy J (2016) The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med 8:595–608. doi:10.15252/emmm.201606210
Shabalin AA (2012) Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28:1353–1358. doi:10.1093/bioinformatics/bts163
Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, Blennow K, Soares H, Simon A, Lewczuk P et al (2009) Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol 65:403–413. doi:10.1002/ana.21610
Skol AD, Scott LJ, Abecasis GR, Boehnke M (2006) Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet 38:209–213. doi:10.1038/ng1706
Sleegers K, Bettens K, De Roeck A, Van Cauwenberghe C, Cuyvers E, Verheijen J, Struyfs H, Van Dongen J, Vermeulen S, Engelborghs S et al (2015) A 22-single nucleotide polymorphism Alzheimer’s disease risk score correlates with family history, onset age, and cerebrospinal fluid Aβ42. Alzheimers Dement 11:1452–1460. doi:10.1016/j.jalz.2015.02.013
Sunderland T, Linker G, Mirza N, Putnam KT, Friedman DL, Kimmel LH, Bergeson J, Manetti GJ, Zimmermann M, Tang B et al (2003) Decreased β-amyloid1-42 and increased tau levels in cerebrospinal fluid of patients with Alzheimer disease. JAMA 289:2094–2103. doi:10.1001/jama.289.16.2094
Trabzuni D, Ryten M, Walker R, Smith C, Imran S, Ramasamy A, Weale ME, Hardy J (2011) Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. J Neurochem 119:275–282. doi:10.1111/j.1471-4159.2011.07432.x
Turner SD (2014) qqman: an R package for visualizing GWAS results using QQ and manhattan plots. bioRxiv doi:10.1101/005165
Van Eldik LJ, Carrillo MC, Cole PE, Feuerbach D, Greenberg BD, Hendrix JA, Kennedy M, Kozauer N, Margolin RA, Molinuevo JL et al (2016) The roles of inflammation and immune mechanisms in Alzheimer’s disease. Alzheimer’s Dement Transl Res Clin Interv 2:99–109. doi:10.1016/j.trci.2016.05.001
Visscher PM, Hemani G, Vinkhuyzen AA, Chen GB, Lee SH, Wray NR, Goddard ME, Yang J (2014) Statistical power to detect genetic (co)variance of complex traits using SNP data in unrelated samples. PLoS Genet 10:e1004269. doi:10.1371/journal.pgen.1004269
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164. doi:10.1093/nar/gkq603
Ward LD, Kellis M (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 40:D930–D934. doi:10.1093/nar/gkr917
Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, Christiansen MW, Fairfax BP, Schramm K, Powell JE et al (2013) Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 45:1238-U1195. doi:10.1038/ng.2756
Willer CJ, Li Y, Abecasis GR (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26:2190–2191. doi:10.1093/bioinformatics/btq340
Yang J, Bakshi A, Zhu Z, Hemani G, Vinkhuyzen AAE, Lee SH, Robinson MR, Perry JRB, Nolte IM, van Vliet-Ostaptchouk JV et al (2015) Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet 47:1114. doi:10.1038/ng.3390
Yang JA, Lee SH, Goddard ME, Visscher PM (2011) GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88:76–82. doi:10.1016/j.ajhg.2010.11.011
Zenaro E, Pietronigro E, Della Bianca V, Piacentino G, Marongiu L, Budui S, Turano E, Rossi B, Angiari S, Dusi S et al (2015) Neutrophils promote Alzheimer’s disease-like pathology and cognitive decline via LFA-1 integrin. Nat Med 21:880–886. doi:10.1038/nm.3913
Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N et al (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34:11929–11947. doi:10.1523/JNEUROSCI.1860-14.2014
Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, Montgomery GW, Goddard ME, Wray NR, Visscher PM et al (2016) Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet 48:481–487. doi:10.1038/ng.3538
Acknowledgements
We thank all the participants and their families, as well as the many institutions and their staff that provided support for all studies involved in this collaboration. We also thank the Alzheimer Disease Genetic Consortium (ADGC) for genotyping and providing data for the BIOCARD, UPENN, HB, SWEDEN, and MAYO cohorts. We thank the Cardiogenics (European Project reference LSHM-CT-2006-037593) project for providing data for the eQTL analysis. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Author contributions
YD analyzed data and wrote the manuscript. ZL verified imputation with genotyped data. MK performed colocalization tests of GWAS and expression data. OH contributed conceptually to the analysis. KB performed genotyping for imputation verification. JLD-A performed disease progression analysis. DC, YC, MVF, JB, SM, BS, BH, KH, and SB prepared genetic data: performed imputation, cleaning, and calculated principal components. AMF, DMH, JCM, SK, AJS., PLDJ., MA, AM, RO, MR, RCP, KB, HZ, LM, VMVD, VM-YL, LMS, JQT, JLH, RM, MAP-V, LAF, ERP, GL, AFDN, ADNI, ADGC, JK and AG provided data. CC prepared the manuscript and supervised the project. All authors read and approved the manuscript.
Author information
Authors and Affiliations
Author notes
J. S. K. Kauwe, A. M. Goate and C. Cruchaga equally contributed to this work.
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Consortia
Corresponding author
Ethics declarations
Conflict of interest
KB and HZ are co-founders of Brain Biomarker Solutions in Gothenburg AB, a GU Venture-based platform company at the University of Gothenburg, Sweden. A.M.G. serves on the SAB for Denali Therapeutics and is the inventor on a patent for MAPT mutations.
Funding
This work was supported by grants from the National Institutes of Health (R01AG044546, P01AG003991, RF1AG053303, R01AG035083, and R01NS085419), and the Alzheimer’s Association (NIRG-11-200110). This research was conducted while C.C. was a recipient of a New Investigator Award in Alzheimer’s disease from the American Federation for Aging Research. C.C. is a recipient of a BrightFocus Foundation Alzheimer’s Disease Research Grant (A2013359S). The recruitment and clinical characterization of research participants at Washington University were supported by NIH P50 AG05681, P01 AG03991, and P01 AG026276. ADGC is supported by grants from the NIH (#U01AG032984) and GERAD from the Wellcome Trust (GR082604MA) and the Medical Research Council (G0300429); additional support was provided by NCRAD (U24 AG21886), NACC (U01 AG016976), NIAGADS (U24-AG041689) and UPENN (P30 AG010124). Support for A.S. was provided by U01 AG024904, R01 AG19771, and P30 AG10133. P.D.J. received support from R01 AG048015. UW ADRC received funding from AG05136. S.K. received support from NIA R03AG050856, Alzheimer’s Association, Michael J. Fox Foundation, and ARUK Biomarkers Across Neurodegenerative Diseases (BAND). M.R. received support from the German Federal Ministry of Education and Research (BMBF) National Genome Research Network (NGFN) Grant No. 01GS08125 and through the Helmholtz Alliance for Mental Health in an Aging Society (HELMA) Grant No. Ha-15. This work was supported by access to equipment made possible by the Hope Center for Neurological Disorders and the Departments of Neurology and Psychiatry at Washington University School of Medicine.
Additional information
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Deming, Y., Li, Z., Kapoor, M. et al. Genome-wide association study identifies four novel loci associated with Alzheimer’s endophenotypes and disease modifiers. Acta Neuropathol 133, 839–856 (2017). https://doi.org/10.1007/s00401-017-1685-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-017-1685-y