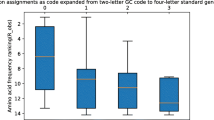
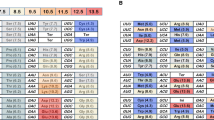
Abstract
There have been two distinct phases of evolution of the genetic code: an ancient phase—prior to the divergence of the three domains of life, during which the standard genetic code was established—and a modern phase, in which many alternative codes have arisen in specific groups of genomes that differ only slightly from the standard code. Here we discuss the factors that are most important in these two phases, and we argue that these are substantially different. In the modern phase, changes are driven by chance events such as tRNA gene deletions and codon disappearance events. Selection acts as a barrier to prevent changes in the code. In contrast, in the ancient phase, selection for increased diversity of amino acids in the code can be a driving force for addition of new amino acids. The pathway of code evolution is constrained by avoiding disruption of genes that are already encoded by earlier versions of the code. The current arrangement of the standard code suggests that it evolved from a four-column code in which Gly, Ala, Asp, and Val were the earliest encoded amino acids.




Similar content being viewed by others
References
Amend JP, Shock EL (1998) Energetics of amino acid synthesis in hydrothermal ecosystems. Science 281:1659–1662
Ardell DH, Sella G (2001) On the evolution of redundancy in genetic codes. J Mol Evol 53:269–281
Bandhu AV, Aggarwal N, Sengupta S (2013) Revisiting the physico-chemical hypothesis of code origin: an analysis based on code-sequence coevolution in a finite population. Orig Life Evol Biosph 43:465–489
Bender A, Hajieva P, Moosmann B (2008) Adaptive antioxidant methionine accumulation in respiratory chain complexes explains the use of a deviant genetic code in mitochondria. Proc Natl Acad Sci USA 105:16496–16501
Bernhardt HS (2012) The RNA world hypothesis: the worst theory of the early evolution of life (except for all the others). Biol Direct 7:23
Burton AS, Lehman N (2009) DNA before proteins? Recent discoveries in nucleic acid catalysis strengthen the case. Astrobiology 9:125–130
Caetano-Anollés G, Seufferheld MJ (2013) The coevolutionary roots of biochemistry and cellular organization challenge the RNA world paradigm. J Mol Microbiol Biotechnol 23:152–177
Cantara WA, Murphy FV, Demirci H, Agris PF (2013) Expanded use of sense codons is regulated by modified cytidines in tRNA. Proc Natl Acad Sci USA 110:10964–10969
Carter CW (2015) What RNA world? Why a peptide/RNA partnership merits renewed experimental attention. Life 5:294–320
Castresana J, Feldmaier-Fuchs G, Pääbo S (1998) Codon reassignment and amino acid composition in hemichordate mitochondria. Proc Natl Acad Sci USA 95:3703–3707
Cleaves HJ (2010) The origin of the biologically coded amino acids. J Theor Biol 263:490–498
Cobb AK, Pudritz RE (2014) Nature’s starships. I. Observed abundances and relative frequencies of amino acids in meteorites. Astrophys J 783:140
Cochella L, Brunelle JL, Green R (2007) Mutational analysis reveals two independent molecular requirements during transfer RNA selection on the ribosome. Nat Struct Mol Biol 14:30–36
Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G (2012) A new understanding of the decoding principle on the ribosome. Nature 484:256–259
Di Giulio M (2002) Genetic code origin: are the pathways of type Glu-tRNA(Gln) –> Gln-tRNA(Gln) molecular fossils or not? J Mol Evol 55:616–622
Di Giulio M (2008) An extension of the coevolution theory of the origin of the genetic code. Biol Direct 3:37
Di Giulio M, Medugno M (1999) Physicochemical optimization in the genetic code origin as the number of codified amino acids increases. J Mol Evol 49:1–10
Di Gulio M (2001) A blind empiricism against the coevolution theory of the origin of the genetic code. J Mol Evol 53:724–732
Francis BR (2011) An alternative to the RNA world hypothesis. Trends Evol Biol 3:e2
Francis BR (2013) Evolution of the genetic code by incorporation of amino acids that improved or changed protein function. J Mol Evol 77:134–158
Francis BR (2015) The hypothesis that the genetic code originated in coupled synthesis of proteins and the evolutionary predecessors of nucleic acids in primitive cells. Life 5:467–505
Freeland SJ, Hurst LD (1998) The genetic code is one in a million. J Mol Evol 47:238–248
Freeland SJ, Wu T, Keulmann N (2003) The case for an error minimizing standard genetic code. Orig Life Evol Biosph 33:457–477
Gilis D, Massar S, Cerf NJ, Rooman M (2001) Optimality of the genetic code with respect to protein stability and amino acid frequencies. Genome Biol 2:11
Goodarzi H, Nejad HA, Torabi N (2004) On the optimality of the genetic code, with the consideration of termination codons. Biosystems 77:163–173
Goodarzi H, Najafabadi HS, Hassani K, Nejad HA, Torabi N (2005) On the optimality of the genetic code, with the consideration of coevolution theory by comparison of prominent cost measure matrices. J Theor Biol 235:318–325
Gromadski KB, Rodnina MV (2004) Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol Cell 13:191–200
Gromadski KB, Daviter T, Rodnina MV (2006) A uniform response to mismatches in codon-anticodon complexes ensures ribosomal fidelity. Mol Cell 21:369–377
Grosjean H, de Crécy-Lagard V, Marck C (2010) Deciphering synonymous codons in the three domains of life: co-evolution with specific tRNA modification enzymes. FEBS Lett 584:252–264
Higgs PG (1998) Compensatory neutral mutations and the evolution of RNA. Genetica 102–103:91–101
Higgs PG (2009) A four-column theory for the origin of the genetic code: tracing the evolutionary pathways that gave rise to an optimized code. Biol Direct 4:16
Higgs PG, Lehman N (2015) The RNA world: molecular co-operation at the origin of life. Nat Rev Genet 16:7–17
Higgs PG, Pudritz RE (2007) From protoplanetary disks to prebiotic amino acids and the origin of the genetic code. In: Pudritz RE, Higgs PG, Stone J (eds) Planetary systems and the origins of life. Cambridge Series in Astrobiology, vol 3. Cambridge University Press, Cambridge
Higgs PG, Pudritz RE (2009) A thermodynamic basis for prebiotic amino acid synthesis and the nature of the first genetic code. Astrobiology 9:483–490
Ibba M, Curnow AW, Söll D (1997) Aminoacyl-tRNA synthesis: divergent routes to a common goal. Trends Biochem Sci 22:39–42
Ibba M, Becker HD, Stathopoulos C, Tumbula DL, Söll D (2000) The adaptor hypothesis revisited. Trends Biochem Sci 25:311–316
Jia W, Higgs PG (2008) Codon usage in mitochondrial genomes: distinguishing context-dependent mutation from translational selection. Mol Biol Evol 25:339–351
Joyce GF (2000) The antiquity of RNA-based evolution. Nature 418:214–221
Jukes TH (1973) Arginine as an evolutionary intruder into protein synthesis. Biochem Biophys Res Commun 53:709–714
Kavran JM, Gundllapalli S, O’Donoghue P, Englert M, Söll D, Steitz TA (2007) Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc Natl Acad Sci USA 104:11268–11273
Knight RD, Landweber LF (2000) Guilt by association: the arginine case revisited. RNA 6:499–510
Knight RD, Freeland SJ, Landweber LF (2001) Rewiring the keyboard: evolvability of the genetic code. Nat Rev Genet 2:49–58
Lajoie MJ, Rovner AJ, Goodman DB, Aerni HR, Haimovich AD, Kuznetsov G, Mercer JA, Wang HH, Carr PA, Mosberg JA, Rohland N, Schultz PG, Jacobson JM, Rinehart J, Church GM, Isaacs FJ (2013) Genomically recoded organisms expand biological functions. Science 342:357–360
Lazcano A, Guerrero R, Margulis L, Oró J (1988) The evolutionary transition from RNA to DNA in early cells. J Mol Evol 27:283–290
Lehman N, Jukes TH (1988) Genetic code development by stop codon takeover. J Theor Biol 135:203–214
Ling J, Daoud R, Lajoie MJ, Church GM, Söll D, Lang BF (2014) Natural reassignment of CUU and CUA sense codons to alanine in Ashbya mitochondria. Nucleic Acids Res 42:499–508
Lozupone CA, Knight RD, Landweber LF (2001) The molecular basis of nuclear genetic code change in ciliates. Curr Biol 11:65–74
Lu Y, Freeland S (2006) On the evolution of the standard amino-acid alphabet. Genome Biol 7:102
Lynch M, Koskella B, Schaack S (2006) Mutation pressure and the evolution of organelle genomic architecture. Science 311:1727–1730
Massey SE, Moura G, Beltrão P, Almeida R, Garey JR, Tuite MF, Santos MA (2003) Comparative evolutionary genomics unveils the molecular mechanism of reassignment of the CTG codon in Candida spp. Genome Res 13:544–557
McCutcheon JP, McDonald BR, Moran NA (2009) Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet 5:e1000565
Miller SL, Cleaves HJ (2007) Prebiotic chemistry on the primitive earth. Orig Life 1:3–56
Miller SL, Urey HC, Oró J (1976) Origin of organic compounds on the primitive earth and in meteorites. J Mol Evol 9:59–72
Muramatsu T, Yokoyama S, Horie N, Matsuda A, Yamaizumi Z, Kuchino Y, Nishimura S, Miyazawa T (1988) A novel lysine-substituted nucleoside in the first position of the anticodon of minor isoleucine tRNA from Escherichia coli. J Biol Chem 263:9261–9267
Novozhilov AS, Koonin EV (2009) Exceptional error minimization in putative primordial genetic codes. Biol Direct 4:44
Osawa S, Jukes TH (1989) Codon reassignment (codon capture) in evolution. J Mol Evol 28:271–278
Osawa S, Collins D, Ohama T, Jukes TH, Watanabe K (1990) Evolution of the mitochondrial genetic code III. Reassignment of CUN codons from leucine to threonine during evolution of yeast mitochondria. J Mol Evol 30:322–328
Pape T, Wintermeyer W, Rodnina M (1999) Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J 18:3800–3807
Philip GK, Freeland SJ (2011) Did evolution select a nonrandom “alphabet” of amino acids? Astrobiology 11:235–240
Ran W, Higgs PG (2010) The influence of anticodon-codon interactions and modified bases on codon usage bias in bacteria. Mol Biol Evol 27:2129–2140
Ronneberg TA, Landweber LF, Freeland SJ (2000) Testing a biosynthetic theory of the genetic code: fact or artifact? Proc Natl Acad Sci USA 97:13690–13695
Rovner AJ, Haimovich AD, Katz SR, Li Z, Grome MW, Gassaway BM, Amiram M, Patel JR, Gallagher RR, Rinehart J, Isaacs FJ (2015) Recoded organisms engineered to depend on synthetic amino acids. Nature 518:89–93
Santos MA, Moura G, Massey SE, Tuite MF (2004) Driving change: the evolution of alternative genetic codes. Trends Genet 20:95–102
Schultz DW, Yarus M (1994) Transfer RNA mutation and the malleability of the genetic code. J Mol Biol 235:1377–1380
Schultz DW, Yarus M (1996) On malleability in the genetic code. J Mol Evol 42:597–601
Sella G, Ardell DH (2002) The impact of message mutation on the fitness of a genetic code. J Mol Evol 54:638–651
Sengupta S, Higgs PG (2005) A unified model of codon reassignment in alternative genetic codes. Genetics 170:831–840
Sengupta S, Yang X, Higgs PG (2007) The mechanisms of codon reassignments in mitochondrial genetic codes. J Mol Evol 64:662–688
Sengupta S, Aggarwal N, Bandhu AV (2014) Two perspectives on the origin of the standard genetic code. Orig Life Evol Biosph 44:287–291
Sheppard K, Yuan J, Hohn MJ, Jester B, Devine KM, Söll D (2008) From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res 36:1813–1825
Su D, Lieberman A, Lang BF, Simonovic M, Söll D, Ling J (2011) An unusual tRNAThr derived from tRNAHis reassigns in yeast mitochondria the CUN codons to threonine. Nucleic Acids Res 39:4866–4874
Suzuki T, Ueda T, Watanabe K (1997) The “polysemous” codon—a codon with multiple amino acid assignment caused by dual specificity of tRNA identity. EMBO J 16:1122–1134
Swire J, Judson OP, Burt A (2005) Mitochondrial genetic codes evolve to match amino acid requirements of proteins. J Mol Evol 60:128–139
Tomita K, Ueda T, Ishiwa S, Crain PF, McCloskey JA, Watanabe K (1999a) Codon reading patterns in Drosophila melanogaster mitochondria based on their tRNA sequences: a unique wobble rule in animal mitochondria. Nucleic Acids Res 27:4291–4297
Tomita K, Ueda T, Watanabe K (1999b) The presence of pseudouridine in the anticodon alters the genetic code: a possible mechanism for assignment of the AAA lysine codon as asparagine in echinoderm mitochondria. Nucleic Acids Res 27:1683–1689
Trifonov EN (2004) The triplet code from first principles. J Biomol Struct Dyn 22:1–11
Tumbula DL, Becker HD, Chang WZ, Söll D (2000) Domain-specific recruitment of amide amino acids for protein synthesis. Nature 407:106–110
Turanov AA, Lobanov AV, Fomenko DE, Morrison HG, Sogin ML, Klobutcher LA, Hatfield DL, Gladyshev VN (2009) Genetic code supports targeted insertion of two amino acids by one codon. Science 323:259–261
van der Gulik PT, Hoff WD (2011) Unassigned codons, nonsense suppression, and anticodon modifications in the evolution of the genetic code. J Mol Evol 73:59–69
Vetsigian K, Woese C, Goldenfeld N (2006) Collective evolution and the genetic code. Proc Natl Acad Sci USA 103:10696–10701
Voorhees RM, Mandal D, Neubauer C, Köhrer C, RajBhandary UL, Ramakrishnan V (2013) The structural basis for specific decoding of AUA by isoleucine tRNA on the ribosome. Nat Struct Mol Biol 20:641–643
Weber AL, Miller SL (1981) Reasons for the occurrence of the twenty coded protein amino acids. J Mol Evol 17:273–284
Wong JT (1975) A co-evolution theory of the genetic code. Proc Natl Acad Sci USA 72:1909–1912
Wong JT (2005) Coevolution theory of the genetic code at age thirty. BioEssays 27:416–425
Wong JT (2014) Emergence of life: from functional RNA selection to natural selection and beyond. Front Biosci 19:1117–1150
Yarus M (2000) RNA-ligand chemistry: a testable source for the genetic code. RNA 6:475–484
Yokobori S, Suzuki T, Watanabe K (2001) Genetic code variations in mitochondria: tRNA as a major determinant of genetic code plasticity. J Mol Evol 53:314–326
Zaher HS, Green R (2009a) Quality control by the ribosome following peptide bond formation. Nature 457:161–166
Zaher HS, Green R (2009b) Fidelity at the molecular level: lessons from protein synthesis. Cell 136:746–762
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sengupta, S., Higgs, P.G. Pathways of Genetic Code Evolution in Ancient and Modern Organisms. J Mol Evol 80, 229–243 (2015). https://doi.org/10.1007/s00239-015-9686-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-015-9686-8