Abstract
The origin of the genetic code marked a major transition from a plausible RNA world to the world of DNA and proteins and is an important milestone in our understanding of the origin of life. We examine the efficacy of the physico-chemical hypothesis of code origin by carrying out simulations of code-sequence coevolution in finite populations in stages, leading first to the emergence of ten amino acid code(s) and subsequently to 14 amino acid code(s). We explore two different scenarios of primordial code evolution. In one scenario, competition occurs between populations of equilibrated code-sequence sets while in another scenario; new codes compete with existing codes as they are gradually introduced into the population with a finite probability. In either case, we find that natural selection between competing codes distinguished by differences in the degree of physico-chemical optimization is unable to explain the structure of the standard genetic code. The code whose structure is most consistent with the standard genetic code is often not among the codes that have a high fixation probability. However, we find that the composition of the code population affects the code fixation probability. A physico-chemically optimized code gets fixed with a significantly higher probability if it competes against a set of randomly generated codes. Our results suggest that physico-chemical optimization may not be the sole driving force in ensuring the emergence of the standard genetic code.
Similar content being viewed by others
Introduction
The standard genetic code (SGC) is nearly universal and is estimated to have originated about 3.8 billion years ago prior to the appearance of the Last Universal Common Ancestor (LUCA) of all known living organisms. Hence, an understanding of the processes that led to the origin of the standard genetic code is crucial for our understanding of the origin of life. Attempts to understand the structure of the SGC has a long history. Several theories (Pelc 1965; Pelc and Welton 1966; Dunnill 1966) were proposed to explain the pattern of amino acid association of codons in the SGC on the basis of stereo-chemical affinities between codons and amino acids. Subsequent lack of evidence rendered such theories ineffective. Nevertheless, a few studies provide some support for the stereo-chemical affinity between Arginine and its codons(Jukes 1973; Knight and Landweber 2000) and Leucine and Tyrosine with their respective codons (Yarus 2000).
Proponents of the physico-chemical (adaptive) theory of code evolution suggested that the genetic code evolved to minimize the effect of mutational (Sonneborn 1965; Epstein 1966) and translational errors (Woese 1965; Goldberg and Wittes 1966; Woese 1967; Di Giulio 1989; Ardell 1998). Woese (1965) was the first to highlight the fact that the arrangement of amino acids among codons in the SGC is non-random. He pointed out, by focusing on the polarity property of amino acids, that amino acids in each of the first two columns of the SGC have similar polarities which reduce the effect of translational errors that replaces one amino acid by another in the same column. Crick (1968) argued that the structure of the SGC may have frozen in an error-correcting form because making further changes in the code would be highly deleterious and hence such organisms which undergo codon reassignments would be selected out of the population. The first influential quantitative studies on the non-random organization of amino acids in the genetic code were carried out by Haig and Hurst (1991) and Freeland and Hurst (1998). They showed, by defining an average cost of translational error, that the SGC is optimized to reduce the cost of translational errors relative to many alternative codes where the amino acids are randomly distributed among the 20 codon blocks. Subsequent refinements of the cost function and particularly the amino acid substitution matrix (Gilis et al. 2001; Goodarzi et al. 2004, 2005; Novozhilov et al. 2007; Chechetkin and Lobzin 2009) have confirmed the highly optimized character of the SGC. Novozhilov et al. (2007) have also examined the fitness landscape of code evolution and found that level of optimization depends on whether the fitness of the random codes is comparable to that of the SGC. The effect of horizontal gene transfer (Vetsigian et al. 2006) on the universality and optimality of the genetic code has also been investigated.
Ardell and Sella, in a series of important papers (Ardell and Sella 2001; Sella and Ardell 2002, 2006), explored the effect of code-sequence co-evolution on the structure of the genetic code using a deterministic, population genetic doublet codon model. Their work provided additional verification of the physico-chemical hypothesis and also yielded several new insights into the early evolution of the code starting from a completely ambiguous coding state in which every codon codes for every biologically encoded amino acid with equal probability. They found that codes always froze before redundancy of codon-amino-acid associations could be removed. Moreover, the final set of encoded amino acids did not span the maximal range of amino acid property space suggesting that code may have evolved to select “generalist” amino acids which can perform a variety of functions, rather than “specialized” amino acids, in order to reduce the effect of translational errors. Zhu and Freeland (2006), building on the work of Orr (1998; 2002), have argued that the SGC in addition to being optimized is also designed to enhance the rate of adaptive evolution.
The co-evolution theory, an influential alternative to the physico-chemical theory of code evolution was proposed by Wong (1975, 1976, 1980, 2005) and subsequently refined and championed by Di Giulio and collaborators (Di Giulio 1996; Di Giulio and Medugno 1998, 1999, 2001; Di Giulio 2008). The co-evolution theory suggests that the structure of the SGC was constrained by the metabolic pathways (Taylor and Coates 1989) of amino acid synthesis. They argued that the code initially encoded a small number of early (precursor) amino acids which could be synthesized abiotically in a few steps from non-amino acid precursors using the glycolytic and citric acid cycle. The structure of the code gradually evolved as redundant codon blocks were ceded from precursor to product amino acids which require precursor amino acids for their synthesis. Di Giulio (2002) has pointed out that synthesis of amino acids on tRNAs could facilitate the ceding of codons from precursor to product amino acids. He cites the evidence of Gln and Asn synthesis from Glu and Asp tRNAs in some Bacteria and Archaea, in support for the co-evolution theory. He further argues (Di Giulio 2008) that charging of tRNAs by aminoacyl tRNA synthetases (aaRS) need not have developed in the earliest stages of code evolution and synthesis of amino-acids on tRNAs may have been the alternative method for charging of tRNAs during that epoch. While that may well be true for some standard amino acids like Asn, Gln, Cys, and a few non-standard ones like Sec and fMet, generalizations to other product amino acids is debatable. Nevertheless, biosynthesis of product amino acids on tRNAs remains one of the strongest signatures of the importance of precursor-product relation between amino acids in shaping the structure of the SGC.
An alternative co-evolution theory proposed by Chechetkin (2006) and Delarue (2007) suggests that structure of the code co-evolved along with the ability of tRNA anticodons to recognize specific codons and aaRS so as to reduce the effect of codon ambiguity and translational errors that characterized the code in the early stages of its evolution.
Despite the initial expectation of a “frozen” SGC (Crick 1968), there is substantial evidence (Knight et al. 2001; Sengupta et al. 2007) to suggest that the code is still evolving. Investigations into the mechanisms of codon reassignments (Swire et al. 2005; Sengupta and Higgs 2005; Sengupta et al. 2007) reveal the complexities involved in producing alternative genetic codes via codon reassignments and provide some hints about the difficulty of expanding the amino acid vocabulary of the code. A better understanding of the structural aspects of the translation machinery and the ability to manipulate them may also provide additional clues on the origin and evolution of the code.
Higgs (2009) proposed a four-column theory for the origin of the genetic code by arguing that the code translational machinery could initially only distinguish between codons which differed in the second base position and encoded amino acids that were associated with codons that encoded G at the first position. The latter hypothesis is based on evidence of sequence fossils found in present day genomes that are characterized by an excess of amino acids encoded by GNN (Brooks and Fresco 2003) and particularly GCU codons (Trifonov and Bettecken 1997; Frenkel and Trifonov 2012). The triplet expansion of the GCU codon followed by point mutations in the tandem GCU repeats may have played a role in determining the association between GCU, codons accessible from GCU by point mutations, and the earliest amino acids. According to the Higgs (2009) model, eventually, the code expanded by reassigning codon blocks from early to late amino acids in an error-correcting manner. By defining a cost function for a code encoding less than 20 amino acids, Higgs showed that driving force behind code expansion in the early stages was primarily positive selection for increased functionality and diversity of encoded proteins resulting from an increase in the encoded amino acid alphabet. Novozhilov and Koonin (2009) also used the code-cost function to draw conclusions about the primordial structure of the genetic code. By making use of the code cost based on polarity values of amino acids to calculate a minimization percentage of a set of codes, they showed that a 2-letter code that distributes ten early amino acids among the sixteen 4-codon blocks in a way that is most consistent with the structure of the SGC also turns out to be the most optimal. Both these studies are based on the premise that code-cost ultimately determines fixation with the most optimal (lowest cost) code guaranteed to be fixed in the population. Such a conclusion is valid only in the limit of infinite population size. Koonin and Novozhilov (2009) provide an excellent review of the many factors that dictated the origin and evolution of the genetic code.
None of the previous literature has addressed the effect of code-sequence co-evolution in finite populations on the early evolution of the genetic code. Finite population effects are particularly important because in such a scenario, fixation of several sub-optimal codes is possible provided their costs are not too different from the most optimal code in the set. That would allow for plausible alternative evolutionary trajectories which eventually lead to the emergence of a universal code. In this paper, we study the effect of code-sequence co-evolution on the evolution of a finite population of primordial codes in the pre-LUCA phase. Our aim is to understand the consequence of competition between a finite population of primordial codes distinguished by differences in the degree of physico-chemical optimization, on the emergence and structure of a universal genetic code. In the process, we aim to explore the conditions under which a population of genetic codes encoding a small number of amino acids are eventually replaced by a set of codes encoding a larger number of amino acids. Even though a genetic code encoding a larger number of amino acids may produce more functionally diverse proteins, the trade-off between the fitness advantage of encoding more amino acids and the disadvantage of changing the code will eventually decide which type of code gets fixed in the population.
The first part of our paper deals with the early phase of code evolution culminating in code(s) which encode ten amino acids. In this stage, we explore two different evolutionary scenarios. In the first scenario, all the plausible codes simultaneously compete with one another. However, the competition starts only after the sequences associated with a code have attained mutation-selection equilibrium. In the second scenario, new codes from the set of plausible codes are gradually and randomly introduced into a population with a fixed probability. In both cases, we compute the probability of fixation of a code. The second part deals with the late stage of code evolution starting from two different initial conditions corresponding to two different ten amino acid codes. In this stage we explore the evolution of codes encoding 10 amino acids to those encoding 14 amino acids. The set of plausible codes selected are constrained by the amino acids available at that stage, the physico-chemical similarity between amino acids encoded in the same column and in some cases the precursor-product relation between amino acids. We also compared the results of code-sequence co-evolution for the set of constrained codes with code-sequence evolution for a set of randomly generated primordial codes not subject to any of the above constraints.
We find that natural selection alone cannot explain the emergence of a single universal code having a structure that is most consistent with the SGC if the pool of competing codes has similar levels of physico-chemical optimization. The structure of the code that gets fixed with highest probability in such a scenario can differ significantly from the one that is most consistent with the SGC, as long as the former satisfies the same physico-chemical constraints in codon amino acid association as that observed in the SGC. Significantly, finite population effects also ensure that slightly sub-optimal codes can get fixed with substantial probability. On the other hand, if a code in the physico-chemically constrained set competes with a set of randomly generated codes with significantly lower levels of physico-chemical optimization, it tends to get fixed with a significantly higher probability than any of the randomly generated (unconstrained) codes.
Model
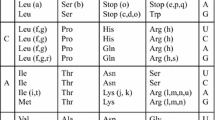
In considering competition between primitive codes, a major challenge lies in identifying a set of plausible primitive codes. The number of possible codes can be prohibitively large. Even if we restrict our choices to those codes that have the same codon block structure as the SGC, the number of possible 20 amino acid codes is 20! (assuming the same amino acid cannot occupy more than one block.) It is unlikely that the pool of competing codes could have been so large. We therefore consider a smaller sub-set of codes which are constrained to distribute the amino acids among the various codon blocks based on certain organizing principles. The physico-chemical similarity between amino acids is one such organizing principle that shaped the structure of the SGC. A Principle Component Analysis (PCA) of amino acids encoded in the SGC shows that amino acids belonging to the first and second columns are tightly clustered. Clustering is observed for the third column amino acids as well, albeit to a lesser extent. This suggests that error minimization brought about by the allocation of similar amino acids in the first column, second column and to a lesser extent in the third column profoundly shaped the primordial evolution of the code. We further hypothesize that the code evolved from one encoding a small number of amino acids by gradually incorporating new amino acids as they were synthesized. Trifonov (2000, 2004) and Higgs and Pudritz (2007, 2009) have established the order of appearance of the 20 biologically encoded amino acids based on an extensive survey of the literature on prebiotic chemistry in which amino acids were synthesized. Their analysis also indicates that the early amino acids required less energy to synthesize and could be easily synthesized from inorganic compounds available in a primordial environment. On the other hand, none of the late amino acids could be synthesized abiotically. The establishment of an early and late amino acid hierarchy is further strengthened by examining the biosynthetic pathways of amino acid synthesis. The co-evolution theory identifies precursor-product relationships between various sets of amino acids. Six of the earliest amino acids can be identified on the basis of the precursor-product classification. Following Trifonov and Higgs & Pudritz, we therefore divide the primitive code evolution process into an early phase that encoded at most the 10 earliest amino acids and a late phase which is marked by the evolution of the code encoding 10 amino acids to a code encoding 14 amino acids. We do not go beyond 14 amino acid codes because subsequent sub-divisions either involve fourth column sub-divisions which do not satisfy the physico-chemical constraints or require distinguishing between purines at the third position, a feature that may likely have appeared quite late in code evolution process.
Our starting point is a four column code proposed by Higgs. In building a set of constrained primitive codes, we assume that code evolution occurred in stages and tRNAs first acquired the ability to distinguish between bases at the second position. This was followed by the gain in ability of the tRNAs to distinguish between bases at the first position and eventually by their ability to distinguish between purines and pyrimidines at the third position. The latter ability leads to sub-division of 4-codon blocks and is incorporated in our model only in the late phase of code evolution. A new code is generated from the 4-column code by reassigning a block of synonymous codons to a new (previously unassigned) amino acid. During the early phase of code evolution, a set of alternative codes are obtained on the basis of the following constraints on reassignments in the first two columns. (i) Codon blocks whose amino acid assignments are consistent with that of the SGC are not reassigned in order to minimize the number of reassignments that would be necessary to attain the SGC from a primitive code. Hence, the GUN and GCN blocks that are associated with amino acids Val and Ala respectively in the 4-column code as well as the SGC do not undergo further reassignments. (ii) A 4-codon block can be reassigned to another amino acid only if it is physico-chemically similar to the original amino acid. Moreover, during the early phase of code evolution, reassignments can occur only within the set of ten earliest amino acids. Consequently, the codon blocks UUN, CUN and AUN can be reassigned from Val to either Leu or Ile but not to Phe or Met since the latter two are late amino acids. Similarly, the codon blocks UCN,CCN,CAN can be reassigned from Ala to either Ser, Pro or Thr; which belong to the set of ten earliest amino acids. (iii) A 4-codon block that has been reassigned once cannot be further reassigned back to its original meaning. It can only be reassigned to a new amino-acid. For example, of the AUN block has been reassigned from Val to Leu once, it cannot be reassigned back from Leu to Val but it can be reassigned from Leu to Ile. Also, reassignments in the first and second columns can at most distinguish between bases at the first position. Hence when such reassignments occur, the entire 4-codon block gets reassigned.
The possible reassignments in the third column are most difficult to predict. This is because the two earliest amino acids in this column, Asp and Glu do not occupy a 4-codon block but partition the lowermost 4-codon block among themselves in the SGC. None of the remaining 4-codon blocks in the SGC are occupied by just one amino acid and all of them are equally partitioned among two amino-acids in the SGC. Moreover, none of the amino acids that appear in the third column, other than Asp and Glu belong to the set of ten earliest amino acids. It therefore seems reasonable to hypothesize that the ability to discriminate between purines and pyrimidines in the third codon position, arose only after the ability to discriminate between purines and pyrimidines at the first codon position. Hence the third column may have been subdivided into 4-codon blocks encoding Asp and Glu in the early phase of code evolution. It is difficult to predict unambiguously which 4-codon block encoded Asp and which Glu. Hence, starting from Asp, we allow for the reassignment of only the upper two 4-codon blocks in the third column to Glu.
The amino acids in the fourth column are least similar in terms of their physicochemical properties. Moreover, apart from Gly and Ser, none of the other fourth column amino acids belong to the set of ten earliest amino acids. Hence, we assume that the fourth column encoded only Gly during the early phase of code evolution.
With the above constraints, the number of alternative codon block assignments in the first column is 2 × 2 × 2 = 8. Similarly, the number of alternative codon block assignments in the second column is 3 × 3 × 3 = 27. The number of alternative codon block assignments in the third column is 1 × 1 = 4 (if we allow for reassignment of YAN from Asp to Glu). Since there are no alternative reassignments possible in the fourth column of the code during the early phase of code evolution, the total number of plausible alternative codes subject to the above constraints is 8 × 27 × 1 = 216. Initially, the frequencies of all codes in the population are taken to be equal.
In building a constrained subset of alternative codes for the late phase of code-sequence coevolution, we assume that the codon block assignments in the first two columns have frozen and the codes differ only in terms of the third column codon assignments. We start from a stage where the third column is subdivided into two eight codon blocks with UAN,CAN being assigned to Glu and AAN,GAN being assigned to Asp. We then build a set of plausible alternative codes obtained by reassignment of 2-codon sub-blocks to appropriate amino acids. In considering plausible reassignments of the 2-codon sub-blocks, we use the following constraints to reduce the number of alternative plausible codes. We first check if physico-chemical similarity exists between Asp (or Glu) and the reassigned amino acids X. We also check if Asp (or Glu) and X shares a precursor-product relationship consistent with co-evolutionary theory. Moreover, if the amino acid associated with a particular 2-codon sub-block is consistent with its assignment in the SGC, then no further reassignments are considered. This constraint is imposed to minimize the number of reassignments that need to be carried out to reach the SGC from a primordial code. Since, GAY is also assigned to Asp in the SGC, we do not consider further reassignment of Asp for that codon sub-block. Finally, if neither physico-chemical similarity, nor precursor-product relationships can explain the amino acid assignment of a third column codon block (example: UAN), then it is not reassigned. In the latter case, the SGC assignment of UAY and UAR are Tyr and Stop respectively. Both Tyr (which is a late amino acid) and Stop were most likely the consequence of very late reassignments and hence we do not consider them. We also do not consider Asp to His reassignment because even though Asp and His are somewhat similar in terms of physico-chemical properties (albeit less so than (Asp, Asn); (Asp, Gln); (Asp, Glu) pairs), they do not share a precursor-product relationship. For the same reason, we do not consider the Glu to Asn reassignment. Furthermore, since third column changes were most likely characterized by the ability of the tRNAs to distinguish between purines and pyrimidines in the third codon position, we consider only those changes which reassign a 2-codon sub-block to a new amino acid X and not consider those changes which reassign an entire 4-codon block to X. In view of these constraints, the plausible reassignments in the third column that lead to alternative genetic codes are: CAY,CAR: Glu to His, Gln, Asp; AAY,AAR: Asp to Asn, Lys, Glu and GAR:Asp to Asn, Lys, Glu. Hence the number of alternative codes that compete with one another in the late phase of code evolution is 108.
We also carried out several simulations of competition between a set of randomly generated codes which included at least one or a few codes belonging to the constrained set generated on the basis of physico-chemical similarity between amino acids. This allowed us to compare the results of competition among the constrained set of codes with that of competition between randomly generated (unconstrained codes) and a code (or a few codes) that satisfied the same physico-chemical constraint observed in the SGC.
The set of primitive codes considered by us is by no means a definitive one. The actual set may have been smaller or somewhat larger. Nevertheless, by imposing the constraint of physico-chemical similarity in obtaining a set of alternative codes, we ensure that the codes considered can be generated by minor changes in the translation machinery which affect codon-amino acid associations. Such changes may have been quite plausible in a primordial world where the translation machinery was still evolving. We therefore feel that our set is representative enough to enable us to address many of the key issues pertaining to code-sequence co-evolution in finite populations.
Methods
In the first scenario, competition occurs between an equilibrated set of code-sequence combinations.
Equilibration Phase
We let a population of 1,000 sequences associated with each code to equilibrate with the corresponding code by allowing each code-sequence set to evolve through mutation and natural selection without requiring them to compete with other code-sequence sets. Initial population consists of identical DNA sequences of length L made up of a combination of sense codons. We consider two different initial sequences in our simulations where each of the sense codons occurs either once (L = 183) or four times (L = 732). In every generation, the sequences undergo random mutations with a mutation rate of μ per site. The number of mutations per sequence is given by a random number chosen from a Poisson distribution with mean μL. A base selected for mutation can mutate to every other base with equal probability following the Jukes-Cantor model. DNA sequences are then translated using their corresponding genetic codes to protein sequences. The fitness of a sequence is calculated by comparing its translated protein sequence with the reference protein sequence which has been obtained by translating the initial sequence with the SGC. The population for the next generation is obtained by successively selecting sequences in the current generation with probabilities proportional to their fitness. This process of mutation, translation and selection continues until a mutation-selection balance is reached and each sequence equilibrates (i.e. mean fitness becomes constant, apart from stochastic fluctuations) with its corresponding genetic code.
Competition Phase
After equilibration, 10 sub-populations each having 100 sequences per code is created by random sampling from the equilibrated populations. Code-sequence sets in each of these 10 sub-populations are made to compete for 10 trials thereby giving a total of 100 trials. This process is repeated for populations equilibrated with 10 different seeds and results of different seeds are combined to get results for a total of 1,000 trials. Sequences following different codes compete with each other for selection to the next generation. This process continues until a code gets fixed in the population. If in Nt trials, a particular genetic code is fixed Nf times then the fixation probability of the code is given by Pf = Nf/Nt. The algorithm for this model is given in Appendix 1.
In the second scenario, we start from an initial equilibrated population of sequences using the five amino acid code which is very similar to the four column code proposed by Higgs but where the third column is equally divided between Asp (AAN,GAN) and Glu (UAN,CAN). We then gradually introduce new codes with a fixed probability per generation by picking a sequence from the population at random and switching the code it uses to translate the sequence to a new one. Unlike the first scenario, the new code-sequence pair starts competing with the existing ones in the population as soon as it is introduced. The gradual introduction of the new code follows two distinct protocols. In the first case, a random code is chosen from the finite set of all possible codes. In the second case, codes are introduced randomly but hierarchically based on the number of amino acids encoded. A new code is randomly selected from a set of codes encoding n-amino acids and competition between two or more n-amino acid codes continues until all n-amino acid codes are introduced and one of them gets fixed in the population. Subsequently, a random code from a set encoding (n + m) amino acids is selected and introduced into the population of n-amino acid codes and allowed to compete with the existing n-amino acid code (m = 1 if n ≥ 7 and m = 2 if n = 5). Thereafter, the process is repeated till the codes with highest number of amino acids have been introduced in the population subsequent to which evolution of the population continues for a fixed number of generations. The simulation is carried out for several trials and the fixation probability is calculated on the basis of those trials where one code gets fixed in the population. Appendix 2 gives the algorithm for the gradual code introduction simulations.
Results
Early Phase of Code Evolution
Competition Between Multiple Equilibrated Code-Sequence Sets
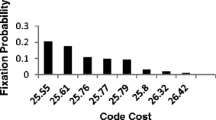
We first discuss the results of competition between 217 codes (including the 4-column code) belonging to the constrained set that is created on the basis of physico-chemical similarity between amino-acids belonging to the same column. Table 1 gives the cost of the ten least costly codes as well as the cost of the 4-column code and the ten amino-acid code that is most consistent with the SGC (labelled CSGC) in terms of amino acid association between codons. The structures of these codes are shown in Fig. 1. The function proposed by Higgs (2009) for calculating the cost of codes encoding less than 20 amino acids is used in the cost-calculation. It is worth noting that CSGC has the 17’th lowest cost but differences in cost between the ten least costly codes is marginal. Table 2 gives the list of codes in the constrained set that has a fixation probability greater than 0.01 and Fig. 2 shows the structure of the ten codes having the highest fixation probability. Figure 3a shows variation in mean fitness of the population after start of competition between codes in one particular trial. Changes in code frequencies for five codes (indicated by different colours) are shown in Fig. 3b. The initial monotonic increase and eventual saturation of the mean fitness can be attributed to the gradual elimination of many low fitness codes and eventual fixation of a single code and establishment of mutation-selection equilibrium between that code and the population of sequences it translates.
Equilibrated code competition model: a Variation of mean fitness of the population with time after the start of competition between 217 codes in the constrained set. b Change in frequency of five codes (indicated by different coloured lines); in one specific trial. Parameters used: No. of sequences per code = 1,000, L = 732, μ = 0.0001, s = 0.05
It is clear that there is no single code which has a significantly higher fixation probability than all other codes. There are at least seven codes with relatively high fixation probabilities (by approximately an order of magnitude). CSGC was fixed only twice out of thousand trials. Sometimes, we also found codes encoding less than ten amino acids that get fixed in the population. Even though in this case, the code with the highest fixation probability also turned out to have the least cost, there were many other codes with comparable fixation probabilities but higher cost. The presence of a large number of codes having similar fixation probabilities can be attributed to the similar levels of optimization of codes belonging to the constrained set. To verify this, we also investigated the effect of competition between 210 randomly generated 10-amino acid codes and CSGC. Here the CSGC emerged as a clear winner and was fixed 539 times out of 1,000 trials, significantly more than any randomly generated code. The cost of the codes in the random set clearly indicates that the CSGC is the most optimized code in the set terms of physico-chemical properties which explains its high fixation probability. We also explored the effect of competition (Table 3) between codes in the random set with CSGC and six other codes having the highest fixation probabilities in the constrained set (see Table 2) to determine the extent to which the composition of code population affects the fixation probability. Here also, the codes which did well in the constrained set were fixed far more frequently and with similar fixation probabilities than either the CSGC (last row in Table 3) or any of the 210 random codes. In an alternative simulation, we determined the result of competition (Table 4) between 210 randomly generated codes, CSGC and six other codes in the constrained set having the least cost (see Table 1). In this case, three of these low cost codes were never fixed in the population. Significantly, the same three codes were also never fixed during the competition between codes belonging to the constrained set. There were four codes (including the CSGC which appears in the second row of Table 4) having similar costs as well as similar fixation probabilities that were significantly larger (more than twice) than other codes in the set. However, these four codes were not the ones with the lowest cost.
We also repeated the simulations for a different parameter set which had significantly larger selection coefficient. Table 5 gives the fixation probabilities for different codes belonging to the constrained set when the length of sequences used is smaller. In this case also, we find many codes to have similar fixation probabilities. As in the previous case, codes with high fixation probabilities do not necessarily have the lowest cost. The CSGC has the 14’th highest fixation probability with a value of 0.031 (not shown in the list) which needs to be contrasted with the highest fixation probability of 0.15. The qualitative trends in variation of mean fitness and code frequency (see Online Resource 1) are similar to that shown in Fig. 3.
Increasing the selection coefficient, which suggests that natural selection should favour more optimized codes, does not seem to significantly increase the fixation probability of low cost codes. This can also be attributed to the fact that many optimized codes have costs that are so similar that even an increase in selection coefficient is incapable of successfully discriminating between these codes.
Competition Between Multiple Code-Sequence Sets in the Gradual Introduction Model
A plausible alternative scenario of the pre-LUCA phase of code evolution may involve the gradual introduction of new codes through reassignment or by conformational changes in the translation machinery. Does this alternative scenario lead to significantly different conclusions for code evolution compared to the previous scenario where all codes are competing with each other simultaneously? In order to explore this question, we carried out simulations where new codes were introduced gradually with a fixed probability per generation starting from an initial state where the entire population consisted of sequences that were translated using the five amino acid code. A new code is introduced when an existing sequence in the population is translated using the new code and has to compete with the rest of the sequences which use different code(s). See “Methods” for details.
Random Introduction of new Codes
It is plausible that alternative primordial codes may have appeared gradually due to variations in the translation machinery and had to compete with an existing set of codes. We therefore explore a model of code evolution in which codes are gradually introduced into the population. The initial state corresponds to an equilibrated population of the 5-amino acid code and new codes are introduced into the population with a fixed probability per generation by randomly picking one code belonging to the finite set of codes. Unlike the previous model, a newly introduced code is not allowed to equilibrate before being forced to compete with other codes in the population. We studied the effect of varying the different model parameters on the fixation probabilities of the codes in the early phase of code evolution. Table 6 shows the fixation probabilities along with the code cost for various values of selection coefficient (s) and new code introduction probability (Pnew). The structures of the top five codes with the highest fixation probabilities for case (b) and (d) are given in Online Resource 2 and Online Resource 3.
While the codes fixed with the highest fixation probabilities are not necessarily the least costly ones for any given sequence length or population size, the fixation probabilities of the codes were much smaller compared to the result of competition between equilibrated code-sequence sets. Many codes got fixed with similar but low fixation probabilities. Since a new code on introduction into the population, does not have the chance to equilibrate before it starts competing with existing code-sequence sets, occasionally the fitness of a new code sequence pair may be high (relative to its equilibrated counterpart) due to the stochastic nature of the switching of a sequence from the old to the new code. This coupled with the random nature of new code introduction and the lower likelihood of slightly fitter variants invading a more adapted population of code(s) can explain the relatively lower fixation probabilities in the gradual introduction model. In this case also, the differences in optimization levels are not significant enough to clearly differentiate between alternative primordial codes in a finite population model. For this reason, we find that eight or nine amino acid codes also get fixed in the population quite frequently. Figure 4 shows the change in mean fitness of the population and the frequency of the different codes that exist in the population at any given time. It is clear from Fig. 4b that most codes that are present in the population have very low frequencies (0.01 or less) and only a few codes exist with significantly higher frequencies, at any given time. The mean fitness initially increases in a step-like manner (unlike the equilibrated code-sequence competition model) and the significant increase in mean fitness around 25,000 generations can be attributed to the appearance and eventual fixation of a fitter code variant. However that code variant is eventually replaced by a code that appears around 63,000 generations. Around 38,000 and 55,000 generations, two new and fitter code variants appear (denoted by grey and yellow lines in Fig. 4b). This is also manifest through a jump in mean fitness around 38,000 generations. However these fitter variants fail to get fixed in the population. Intriguingly, the appearance of a new and less fit code variant (an 8-amino acid code; see Online Resource 4) around 63,000 generations (pink line in Fig. 4b), which eventually gets fixed by invading the existing fitter code variant, is correlated with a dip in mean fitness. Nevertheless, the mean fitness subsequently increases possibly due to mutation-selection equilibrium being established between the code and the sequences it translates. Subsequent new code variants which appear after 70,000 generations cause small fluctuations in the mean fitness but fail to get fixed in the population and are eventually eliminated. For almost all (except one) parameter sets explored, CSGC does not occur among the top ten codes with the highest fixation probability. For the parameter set specified in Table 6(c), the CSGC has the 15’th highest fixation probability (=0.011) which is to be contrasted with the highest fixation probability (=0.037).
Random code introduction model: a Change in mean fitness of the population with time. b Frequency of the different codes (indicated by different coloured lines) in the population; for a single trial. The code indicated by the pink line that appears in the population around 63,000 generations eventually gets fixed. Parameters used: L = 732, N = 1,000, μ = 0.0001, s = 0.02, Pnew = 0.1
For larger populations, the difference between fixation probabilities of alternative codes reduces even further with the ten codes having almost equal and low fixation probabilities. Online Resource 5 shows the code costs for codes with ten highest fixation probabilities for the same parameter set as specified in Fig. 4 but with N = 10000. Online Resource 6 shows the structure of the codes listed in Online Resource 5 and Online Resource 7(a) and (b) shows the change in mean fitness and the code frequency. On comparing Fig. 4b and Online Resource 7, it becomes evident that an increase in population size makes it less likely for a code invasion to occur more than once. When new codes appear, their frequency seldom increases beyond 0.001 for sequences with L = 732. This implies that the fitness advantage associated with a new code must be considerably large for the new code to be able to invade the population. This is manifest in the figures given in Online Resource 7 which shows that the fixation of a new code variant that appears just before 50,000 generations is marked by an increase in mean fitness. Further subsequent increase in mean fitness occurs as the sequences gradually get equilibrated with this new code variant and saturation in the meant fitness occurs after mutation-selection equilibration is established.
Hierarchical Introduction of new Codes
In this scenario new codes are introduced hierarchically (see “Methods” section for details) from the finite set of physico-chemically constrained codes, based on the number of amino acids encoded. Here too, the codes fixed with the highest probability are not the least costly codes for any parameter set. Table 7 gives the fixation probabilities versus code cost for two different values of selection coefficient. The CSGC appears within the top twenty highest fixation probability codes for most of the parameter sets explored. Figure 5a shows the variation in mean fitness and Fig. 5b shows the frequency of the various codes present in the population. A low value of the selection coefficient enables many more codes to survive in the population especially since in this case, codes encoding the same number of amino acids are more likely to compete against each other at any given time. This is evident in Fig. 5b which shows many instances of invasion by new codes as well as coexistence of several codes at any given time. Hence, even low fitness codes to occasionally become fixed in the population. The mean fitness of the population sometimes increases in almost a step-like manner. This happens when a significantly fitter code variant encoding a larger number of amino acids, invades an existing population. This effect is more striking when the selection coefficient is high. (See Online Resource 8).
Hierarchical code introduction model: a Change in mean fitness of the population with time. b Frequency of the different codes (indicated by different coloured lines) in the population; for a single trial. At any generation a few codes are present in the population and a new code invasion is more probable. Parameters used: s = 0.02, Pnew =0.5, μ = 0.0001, N = 1,000, L = 732
The results of the hierarchical code introduction model are also consistent with those described in the previous sub-sections and suggest that it is practically impossible to distinguish between several alternative codes having very similar levels of optimization. How does a physico-chemically optimized code like CSGC or some other code with an even higher level of optimization fare against randomly generated (unconstrained) codes in the gradual code introduction model? In order to address this question, we first generated a set of 210 random codes (encoding 8–10 amino acids) that are not subject to physico-chemical constraints along with CSGC which was the only optimized code in the set. In a simulation where codes were gradually introduced from this set, the CSGC was the clear winner with a fixation probability that was substantially larger than any of the unconstrained codes that had significantly higher costs. A population of unconstrained codes was easily invaded by CSGC. However, this result does not in imply there is anything special about the structure of CSGC. When CSGC was replaced by another code from the constrained set that had a slightly higher code-cost compared to CSGC, the latter was fixed far more frequently than any of the unconstrained codes.
Late Stage of Code Evolution
Competition Between Multiple Equilibrated Code-Sequence Sets
In this stage, we consider the evolution of the code from one which encodes ten amino acids to one which encodes 14 amino acids. We assume that the code assignments in the first two columns have been frozen and code-expansion occurs only through reassignments in the third column. We do not consider further subdivisions of codon blocks in the first column since Phe and especially Met appear very late in the temporal hierarchy of the 20 biologically encoded amino acids. We also do not consider any subdivisions of the fourth column since they are impossible to predict on the basis of either the physico-chemical or the coevolution hypothesis. We consider two ten-amino acid codes as starting points for code evolution in the late phase. These are CSGC and another ten amino acid code that had the highest fixation probability in the early phase of code evolution (see codes labelled “CSGC” in Fig. 1 and code labelled “1” in Fig. 2). The set of constrained codes are obtained on the basis of reassignments that are consistent with either the physico-chemical hypothesis or the co-evolution hypothesis (see “Methods” section for details). Our aim as before is to ascertain whether natural selection between alternative codes belonging to this constrained set can lead to the emergence of a 14 amino acid code whose structure is consistent with that of the SGC (referred to as CSGC14). Table 8(a) and (b) lists the code cost of five codes that got fixed with the highest fixation probability for the starting codes labelled “CSGC” in Fig. 1 and “1” in Fig. 2 respectively. In the former case, even though CSGC14 was found to have the second highest fixation probability of 0.193, there were six codes that had similar fixation probabilities. In the latter case, CSGC14 had a very low fixation probability that was two orders of magnitude lower than the five highest fixation probability codes. The structures of these five codes are given in Online Resource 9 and Online Resource 10.
Conclusions
The finite population dynamics of code-sequence co-evolution presented here provides several insights into the early evolution of the genetic code. Conclusions based on infinite population models of code evolution need to be re-evaluated in the light of our results. Our results suggest that the cost of the code, as measured by the degree of physico-chemical optimization, is not sufficient to determine fixation of the code. We found several codes in our constrained set which have higher cost than the most optimal code and yet got fixed in the population with sufficiently high probability. In both our models and for both phases of code evolution, it was difficult to distinguish between the fifteen to twenty codes from the constrained set, with the highest fixation probabilities. The CSGC was often not among the top ten codes with the highest fixation probabilities. The small differences in fitness between several codes belonging to the constrained set ensured that stochastic fluctuations arising from the finite population size played an important role in code fixation. This leads us to conclude that the code evolution trajectory is possibly affected by extraneous factors and cannot be explained solely by natural selection between competing codes distinguished by differences in the level of physico-chemical optimization. However, as anticipated, a code belonging to the constrained set is fixed with high probability if it competes only with a randomly generated (non-optimized) set of codes. In the language of fitness landscapes, the codes belonging to the constrained set correspond to local peaks in the fitness landscape whose heights are not significantly different.
The code population structure at any given time clearly affects code fixation. This is also evident from the results of our simulations of the late stage of code evolution and the gradual code introduction model. In the former scenario, the initial state of the system defined by the structure of the 10 amino acid code determines the outcome of competition that results in the eventual fixation of a 14 amino acid code. In the latter scenario a non-equilibrated code has a lower likelihood of invading a possibly better adapted population of one or more codes. On the contrary, there is a high probability that such a code will be quickly removed from the population. Hence at any given point of time, only a few competing codes are present in the population. An optimized code with a significant fitness advantage over existing codes in the population can invade the population with a higher probability. However, if it has to compete with code(s) having similar optimization levels, its ability to invade the population will depend approximately inversely on the population size as predicted by neutral evolution theory.
It is worth emphasizing that we do not deny the importance of physico-chemical optimization in shaping the evolution of the code structure. The redundancy of codon-amino acid associations observed in the SGC and physico-chemical similarity of amino acids in the first and second column (and to a lesser extent in the third column) of the SGC clearly suggests that the code evolved to reduce the effect of translational errors. However our results indicate that the selection of the SGC over alternative codes which differ marginally in the degree of optimality cannot be explained only by the physico-chemical hypothesis of code origin. Such alternative codes could easily have emerged due to small changes in the evolving translation machinery in the pre-LUCA phase. The appearance of such alternative codes may have been facilitated by smaller genomes that were most likely prevalent in the pre-LUCA phase. The post-LUCA evolution of the genetic code observed in many mitochondrial genomes (Knight et al. 2001; Sengupta et al. 2007; Swire et al. 2005) lends further support to the plausibility of appearance of organisms following alternative codes with distinct but similar optimization levels.
The structure of the earliest primordial code(s) and the nature of the molecular machinery that was responsible for establishing such a code are also likely to have affected the pathways of code evolution. This is a challenging issue to resolve and lie at the heart of some of the disagreements between the competing theories of code origin. We feel that the code evolution by sub-division of codon blocks could have been driven by a combination of physico-chemical optimization and ceding of codon blocks to new amino acids based on precursor-product relation between old and new amino acids. Sometimes, though not always, sub-divisions based on precursor-product relationships may also have been compatible with constraints imposed by the physico-chemical optimization hypothesis. Hence, the co-evolution theory of code-evolution may not necessarily be incompatible with the adaptive theory, a conclusion also reached by Di-Giulio (2005).
The evolution of the molecular recognition mechanisms that lead to aminoacylation of the tRNA, binding of the tRNA to the ribosome and binding of the tRNA anti-codons with the codons in the mRNA, were also crucial determinants in code evolution. The evolution of the specificity of those reactions most likely tipped the balance in favour of a particular code structure thereby leading to a universal code. The genetic code provides a recipe for protein synthesis only because aminoacylated tRNAs with the appropriate anti-codons pair with the corresponding codons in the mRNA. This process requires a fully developed translational machinery which involves the ribosome, aaRS, initiation factors and release factors. Most studies on genetic code origin assume that the translation machinery was well-developed (albeit, available in several variants) while associating codons with amino acids in various genetic codes. However, this leads to a conundrum which can only be solved when an understanding of the origin of the translation machinery emerges. Wolf and Koonin (2007) have suggested a step-wise method by which the complex molecular machinery responsible for protein synthesis could have evolved progressively towards increasing complexity from functionally simpler components. This process of coevolution of the translation machinery and the genetic code could have been important in ensuring the emergence of the SGC. It appears that Crick’s “frozen accident” hypothesis of code origin may be more prophetic than previously anticipated.
References
Ardell DH (1998) On error minimization in a sequential origin of the standard genetic code. J Mol Evol 47:1–13
Ardell DH, Sella G (2001) On the evolution of redundancy in genetic codes. J Mol Evol 53:269–281
Brooks DJ, Fresco JR (2003) Greater GNN pattern bias in sequence elements encoding conserved residues of ancient proteins may be an indicator of amino acid composition of early proteins. Gene 303:177–185
Chechetkin VR (2006) Genetic code from tRNA point of view. J Theor Biol 242:922–934
Chechetkin VR, Lobzin VV (2009) Local stability and evolution of the genetic code. J Theor Biol 261:643–653
Crick FHC (1968) The origin of the genetic code. J Mol Biol 38:367–379
Delarue M (2007) An asymmetric underlying rule in the assignment of codons: possible clue to a quick early evolution of the genetic code via successive binary choices. RNA 13:161–169
Di Giulio M (1989) The extension reached by the minimization of polarity distances during the evolution of the genetic code. J Mol Evol 29:288–293
Di Giulio M (1996) The β-sheets of proteins, the biosynthetic relationships between amino acids and the origin of the genetic code. Orig Life Evol Biosph 26:589–609
Di Giulio M (2002) Genetic code origin: are the pathways of type Glu-tRNA(Gln) – > Gln-tRNA(Gln) molecular fossils or not? J Mol Evol 55:616–622
Di Giulio M (2005) The origin of the genetic code: theories and their relationships, a review. BioSystems 80:175–184
Di Giulio M (2008) An extension of the coevolution theory of the origin of the genetic code. Biol Direct 3:37
Di Giulio M, Medugno M (1998) The historical factor: The biosynthetic relationships between amino acids and their physicochemical properties in the origin of the genetic code. J Mol Evol 46:615–621
Di Giulio M, Medugno M (1999) Physicochemical optimization in the genetic code origin as the number of codified amino acids increases. J Mol Evol 49:1–10
Di Giulio M, Medugno M (2001) The level and landscape of optimization in the origin of the genetic code. J Mol Evol 52:372–382
Dunnill P (1966) Triplet nucleotide-amino-acid pairing; a stereochemical basis for the division between protein and non-protein amino-acids. Nature 210:1265–1267
Epstein C (1966) Role of the amino-acid “code” and of selection for conformation in the evolution of proteins. Nature 210:25–28
Freeland SJ, Hurst LD (1998) The genetic code is one in a million. J Mol Evol 47:238–248
Frenkel ZM, Trifonov EN (2012) Origin and evolution of genes and genomes. Crucial role of triplet expansions. J Biomol Struct Dyn 30(2):201–210
Gilis D, Massar S, Cerf NJ, Rooman M (2001) Optimality of the genetic code with respect to protein stability and amino-acid frequencies. Genome Biol 2:49.1–49.12
Goldberg AL, Wittes RE (1966) Genetic code: aspects of organization. Science 153:420–424
Goodarzi H, Nejad HA, Torabi N (2004) On the optimality of the genetic code, with the consideration of termination codons. Biosystems 77:163–173
Goodarzi H, Najafabadi HS, Hassani K et al (2005) On the optimality of the genetic code, with the consideration of coevolution theory by comparison of prominent cost measure matrices. J Theor Biol 235:318–325
Haig D, Hurst LD (1991) A quantitative measure of error minimization in the genetic code. J Mol Evol 33:412–417
Higgs PG (2009) A four-column theory for the origin of the genetic code: tracing the evolutionary pathways that gave rise to an optimized code. Biol Direct 4:16
Higgs PG, Pudritz R (2007) From protoplanetary disks to prebiotic amino acids and the origin of the genetic code. Cambridge University Press, Cambridge
Higgs PG, Pudritz RE (2009) A thermodynamic basis for prebiotic amino acid synthesis and the nature of the first genetic code. Astrobiology 9:483–490
Jukes TH (1973) Arginine as an evolutionary intruder into protein synthesis. Biochem Biophys Res Commun 53:709–714
Knight RD, Landweber LF (2000) Guilt by association: the arginine case revisited. RNA 6:499–510
Knight RD, Freeland SJ, Landweber LF (2001) Rewiring the keyboard: evolvability of the genetic code. Nat Rev Genet 2:49–58
Koonin EV, Novozhilov AS (2009) Origin and evolution of the genetic code: the universal enigma. IUBMB Life 61:99–111
Novozhilov AS, Koonin EV (2009) Exceptional error minimization in putative primordial genetic codes. Biol Direct 4:44
Novozhilov AS, Wolf YI, Koonin EV (2007) Evolution of the genetic code: partial optimization of a random code for robustness to translation error in a rugged fitness landscape. Biol Direct 2:24
Orr HA (1998) The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution (N Y) 52:935
Orr HA (2002) The population genetics of adaptation: the adaptation of DNA sequences. Evolution (N Y) 56:1317–1330
Pelc S (1965) Correlation between coding-triplets and amino-acids. Nature 207:597–599
Pelc S, Welton M (1966) Stereochemical relationship between coding triplets and amino-acids. Nature 209:868–870
Sella G, Ardell DH (2002) The impact of message mutation on the fitness of a genetic code. J Mol Evol 54:638–651
Sella G, Ardell DH (2006) The coevolution of genes and genetic codes: Crick’s frozen accident revisited. J Mol Evol 63:297–313
Sengupta S, Higgs PG (2005) A unified model of codon reassignment in alternative genetic codes. Genetics 170:831–840
Sengupta S, Yang X, Higgs PG (2007) The mechanisms of codon reassignments in mitochondrial genetic codes. J Mol Evol 64:662–688
Sonneborn T (1965) Degeneracy of the genetic code: extent, nature, and genetic implications. Evol genes proteins Acad Press New York 377–397
Swire J, Judson OP, Burt A (2005) Mitochondrial genetic codes evolve to match amino acid requirements of proteins. J Mol Evol 60:128–139
Taylor FJR, Coates D (1989) The code within the codons. BioSystems 22:177–187
Trifonov EN (2000) Consensus temporal order of amino acids and the evolution of the triplet code. Gene 261:139–151
Trifonov EN (2004) The triplet code from first principles. J Biomol Struct Dyn 22:1–11
Trifonov EN, Bettecken T (1997) Sequence fossils, triplet expansion, and reconstruction of earliest codons. Gene 205:1–6
Vetsigian K, Woese C, Goldenfeld N (2006) Collective evolution and the genetic code. Proc Natl Acad Sci U S A 103:10696–10701
Woese CR (1965) Order in the genetic code. Proc Natl Acad Sci U S A 54:71–75
Woese CR (1967) The genetic code: the molecular basis for genetic expression. Harper & Row, New York
Wolf YI, Koonin EV (2007) On the origin of the translation system and the genetic code in the RNA world by means of natural selection, exaptation, and subfunctionalization. Biol Direct 2:14
Wong JT (1975) A co-evolution theory of the genetic code. Proc Natl Acad Sci U S A 72:1909–1912
Wong JT (1976) The evolution of a universal genetic code. Proc Natl Acad Sci U S A 73:2336–2340
Wong JT (1980) Role of minimization of chemical distances between amino acids in the evolution of the genetic code. Proc Natl Acad Sci U S A 77:1083–1086
Wong JT (2005) Coevolution theory of the genetic code at age thirty. Bioessays 27:416–425
Yarus M (2000) RNA-ligand chemistry: a testable source for the genetic code. RNA 6:475–484
Zhu W, Freeland S (2006) The standard genetic code enhances adaptive evolution of proteins. J Theor Biol 239:63–70
Acknowledgments
We thank Devapriya Choudhury for valuable discussions. AVB is supported by the Council of Scientific and Industrial Research (CSIR) senior research fellowship and NA is supported by a DBT-BINC fellowship provided by the Department of Biotechnology (DBT), India.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
(PDF 28 kb)
Online Resource 2
(PDF 18 kb)
Online Resource 3
(PDF 18 kb)
Online Resource 4
(PDF 200 KB)
Online Resource 5
(PDF 23 kb)
Online Resource 6
(PDF 27 kb)
Online Resource 7
(PDF 23 kb)
Online Resource 8
(PDF 26 kb)
Online Resource 9
(PDF 19 kb)
Online Resource 10
(PDF 19 kb)
Appendices
Appendix 1
We give below the algorithm for code-sequence coevolution for the case where the sequences are first equilibrated to their respective codes and all the equilibrated set of code-sequence combinations compete against each other simultaneously.
-
1)
Start with a population containing 1,000 identical nucleotide sequences of length L corresponding to each code.
-
2)
Each code-sequence set is allowed to evolve through mutation and natural selection without requiring it to compete with other code-sequence sets till sequences have adapted to their respective genetic codes and mutation-selection equilibrium is established.
-
3)
Mutations are made in the sequences in accordance with the Jukes-Cantor model, with a mutation rate of μ. The mutation-selection process is carried out as follows:
-
i.
Copy the parent sequence to the offspring sequence.
-
ii.
Make mutations in the offspring sequences with the number of mutations, n determined by the Poisson distribution.
$$ p(n)=\frac{e{\hbox{--}}^{\mu L}{\left(\mu L\right)}^n}{n!} $$ -
iii.
Choose the position in the sequence that is to be mutated from a uniform distribution between 1 and L.
-
iv.
Make mutations according to Jukes-Cantor mutation matrix.
-
i.
-
4)
Compute the fitness of each offspring sequence by translating the sequence with the code it follows and comparing the generated amino acid sequence with the reference amino acid sequence obtained by translating the initial sequence with the standard genetic code. We assume that the fitness of a sequence is given by the product of the fitness of individual codons making up the sequence. This implies the absence of any correlations between codons making up the sequence. Hence the fitness of the i’th sequence is given by
$$ w(i)={\displaystyle \prod_{k=1}^{L/3}\left(1- sg\left({a}_k,{b}_k\right)\right)} $$where, a k is the amino acid associated with the k’th codon in the reference protein sequence; b k is the corresponding amino acid as specified in the code associated with the evolving sequence and g(a.b) is the cost of replacing amino acid a by amino acid b. g(a,b) has been normalized so that its maximum possible value is 1. s is the selection coefficient.
-
5)
The parent population for the next generation (t + 1) is obtained by selecting sequences in the offspring population in the current generation (t) with probabilities proportional to their fitness.
-
6)
The equilibrated population is then used for creating 10 sub-populations. Every sub-population has 100 sequences (corresponding to each of the genetic codes)that are randomly selected from the set of 1,000 equilibrated sequences.
-
7)
Code-sequence sets in each of these 10 sub-populations are made to compete against each other until a code gets fixed in the population and this process is repeated for 10 trials. The results of each sub-population are combined to generate results for 100 trials.
-
8)
Steps 6 and 7 are repeated for populations equilibrated with ten different random number seeds and results thus obtained are combined to get results for a total of 1,000 trials.
-
9)
If in Nt trials a particular genetic code is fixed Nf times, the fixation probability of the code is given by Pf = Nf / Nt
Appendix 2
We give below the algorithm for code-sequence coevolution for the case where codes are gradually introduced into the population. In the first scenario, a new code is introduced by randomly selecting one from a pre-defined set of codes.
-
1.
Start with a population of N sequences of length L each, following the five amino acid code.
-
2.
The sequences are allowed to evolve and reach mutation-selection equilibrium with respect to the five amino acid code by following steps (3)–(5) in Appendix 1 till the mean fitness of the population becomes constant (apart from stochastic fluctuations).
-
3.
New codes are introduced in the population with a fixed probability. For this, a random number between 1 and N is generated to select the sequence which is translated with the new code.
-
4.
The population is evolved by repeating steps (3)–(5) of Appendix 1 and the simulation is run for a fixed number of generations to identify the code which has the maximum frequency in the last generation or until one of the codes become fixed in the population, whichever occurs earlier.
-
5.
Steps (2)–(4) are then repeated for a fixed number of trials (Nt) and the fixation probability is calculated on the basis of those trials where a code gets fixed in the population.
For the second scenario (hierarchical code introduction model) the step (3) in the above algorithm was modified as follows:
-
i.
The codes are introduced in the population with a finite probability from a set of codes encoding the least number (n) of amino acids. The population is allowed to evolve by mutation and selection till all the codes from the current set of n amino acid codes have been introduced and one of them gets fixed in the population.
-
ii.
Thereafter, the codes to be introduced are chosen from a set encoding the next highest number of amino acids and this process is repeated till all the codes from the set of codes encoding highest number of amino acids (i.e. 10) have been introduced in the population.
Rights and permissions
About this article
Cite this article
Bandhu, A.V., Aggarwal, N. & Sengupta, S. Revisiting the Physico-Chemical Hypothesis of Code Origin: An Analysis Based on Code-Sequence Coevolution in a Finite Population. Orig Life Evol Biosph 43, 465–489 (2013). https://doi.org/10.1007/s11084-014-9353-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11084-014-9353-x