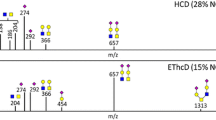
Abstract
Growing evidence on the diverse biological roles of extracellular glycosylation as well as the need for quality control of protein pharmaceuticals make glycopeptide analysis both exciting and important again after a long hiatus. High-throughput O-glycosylation studies have to tackle the complexity of glycosylation as well as technical difficulties and, up to now, have yielded only limited results mostly from single enrichment experiments. In this study, we address the technical reproducibility of the characterization of the most prevalent O-glycosylation (mucin-type core 1 structures) in human serum, using a two-step lectin affinity-based workflow. Our results are based on automated glycopeptide identifications from higher-energy C-trap dissociation and electron transfer dissociation MS/MS data. Assignments meeting strict acceptance criteria served as the foundation for generating “spectral families” incorporating low-scoring MS/MS identifications, supported by accurate mass measurements and expected chromatographic retention times. We show that this approach helped to evaluate the reproducibility of the glycopeptide enrichment more reliably and also contributed to the expansion of the glycoform repertoire of already identified glycosylated sequences. The roadblocks hindering more in-depth investigations and quantitative analyses will also be discussed.






Similar content being viewed by others
References
Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–49.
Varki A. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology. 1993;3:97–130.
Haltiwanger RS, Lowe JB. Role of glycosylation in development. Annu Rev Biochem. 2004;73:491–537.
Rudd PM, Elliott T, Cresswell P, Wilson IA, Dwek RA. Glycosylation and the immune system. Science. 2001;291:2370–76.
Brockhausen I. Mucin-type O-glycans in human colon and breast cancer: glycodynamics and functions. EMBO Rep. 2006;7:599–604.
Arnold JN, Saldova R, Hamid UM, Rudd PM. Evaluation of the serum N-linked glycome for the diagnosis of cancer and chronic inflammation. Proteomics. 2008;8:3284–93.
Grigorian A, Mkhikian H, Li CF, Newton BL, Zhou RW, Demetriou M. Pathogenesis of multiple sclerosis via environmental and genetic dysregulation of Nglycosylation. Semin Immunopathol. 2012;34:415–24.
Christiansen MN, Chik J, Lee L, Anugraham M, Abrahams JL, Packer NH. Cell surface protein glycosylation in cancer. Proteomics. 2013;14:525–46.
Scott DW, Patel RP. Endothelial heterogeneity and adhesion molecules N-glycosylation: implications in leukocyte trafficking in inflammation. Glycobiology. 2013;23:622–33.
Stuchlova Horynova M, Raska M, Clausen H, Novak J. Aberrant O-glycosylation and anti-glycan antibodies in an autoimmune disease IgA nephropathy and breast adenocarcinoma. Cell Mol Life Sci. 2013;70:829–39.
Venkatakrishnan V, Packer NH, Thaysen-Andersen M. Host mucin glycosylation plays a role in bacterial adhesion in lungs of individuals with cystic fibrosis. Expert Rev Respir Med. 2013;7:553–76.
Schedin-Weiss S, Winblad B, Tjernberg LO. The role of protein glycosylation in Alzheimer disease. FEBS J. 2014;281:46–62.
Mehta A, Comunale MA, Rawat S, Casciano JC, Lamontagne J, Herrera H, et al. Intrinsic hepatocyte dedifferentiation is accompanied by upregulation of mesenchymal markers, protein sialylation and core alpha 1,6 linked fucosylation. Sci Rep. 2016;6:27965.
Wi GR, Moon BI, Kim HJ, Lim W, Lee A, Lee JW, et al. A lectin-based approach to detecting carcinogenesis in breast tissue. Oncol Lett. 2016;11:3889–95.
Strum JS, Nwosu CC, Hua S, Kronewitter SR, Seipert RR, Bachelor RJ, et al. Automated assignments of N- and O-site specific glycosylation with extensive glycan heterogeneity of glycoprotein mixtures. Anal Chem. 2013;85:5666–75.
Hoffmann M, Marx K, Reichl U, Wuhrer M, Rapp E. Site-specific O-glycosylation analysis of human blood plasma proteins. Mol Cell Proteomics. 2016;15:624–41.
Hägglund P, Matthiesen R, Elortza F, Højrup P, Roepstorff P, Jensen ON, et al. An enzymatic deglycosylation scheme enabling identification of core fucosylated N-glycans and O-glycosylation site mapping of human plasma proteins. J Proteome Res. 2007;6:3021–31.
Alves MJ, Kawahara R, Viner R, Colli W, Mattos EC, Thaysen-Andersen M, et al. Comprehensive glycoprofiling of the epimastigote and trypomastigote stages of Trypanosoma cruzi. J Proteomics. 2016. doi:10.1016/j.jprot.2016.05.034.
Nilsson J, Rüetschi U, Halim A, Hesse C, Carlsohn E, Brinkmalm G, et al. Enrichment of glycopeptides for glycan structure and attachment site identification. Nat Methods. 2009;6:809–11.
Halim A, Nilsson J, Rüetschi U, Hesse C, Larson G. Human urinary glycoproteomics; attachment site specific analysis of N- and O-linked glycosylations by CID and ECD. Mol Cell Proteomics. 2012. doi:10.1074/mcp.M111.013649.
Halim A, Rüetschi U, Larson G, Nilsson J. LC-MS/MS characterization of O-glycosylation sites and glycan structures of human cerebrospinal fluid glycoproteins. J Proteome Res. 2013;12:573–84.
Darula Z, Medzihradszky KF. Affinity enrichment and characterization of mucin core-1 type glycopeptides from bovine serum. Mol Cell Proteomics. 2009;8:2515–26.
Darula Z, Sherman J, Medzihradszky KF. How to dig deeper? Improved enrichment methods for mucin core-1 type glycopeptides. Mol Cell Proteomics. 2012. doi:10.1074/mcp.O111.016774.
Darula Z, Sarnyai F, Medzihradszky KF. O-glycosylation sites identified from mucin core-1 type glycopeptides from human serum. Glycoconj J. 2016;33:435–45.
Bai X, Li D, Zhu J, Guan Y, Zhang Q, Chi L. From individual proteins to proteomic samples: characterization of O-glycosylation sites in human chorionic gonadotropin and human-plasma proteins. Anal Bioanal Chem. 2015;407:1857–69.
Trinidad JC, Schoepfer R, Burlingame AL, Medzihradszky KF. N- and O-glycosylation in the murine synaptosome. Mol Cell Proteomics. 2013. doi:10.1074/mcp.M113.030007.
Medzihradszky KF, Kaasik K, Chalkley RJ. Tissue-specific glycosylation at the glycopeptide level. Mol Cell Proteomics. 2015. doi:10.1074/mcp.M115.050393.
Medzihradszky KF, Kaasik K, Chalkley RJ. Characterizing sialic acid variants at the glycopeptide level. Anal Chem. 2015;87:3064–71.
Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, et al. Electron capture dissociation for structural characterization of multiply charged protein cations. Anal Chem. 2000;72:563–73.
Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101:9528–33.
Alpert AJ. Electrostatic repulsion hydrophilic interaction chromatography for isocratic separation of charged solutes and selective isolation of phosphopeptides. Anal Chem. 2008;80:62–76.
Chang CF, Pan JF, Lin CN, Wu IL, Wong CH, Lin CH. Rapid characterization of sugar-binding specificity by in-solution proximity binding with photosensitizers. Glycobiology. 2011;21:895–902.
Hortin GL. Isolation of glycopeptides containing O-linked oligosaccharides by lectin affinity chromatography on Jacalin-agarose. Anal Biochem. 1990;191:262–7.
Durham M, Regnier FE. Targeted glycoproteomics: serial lectin affinity chromatography in the selection of O-glycosylation sites on proteins from the human blood proteome. J Chromatogr A. 2006;1132:165–73.
Yabu M, Korekane H, Miyamoto Y. Precise structural analysis of O-linked oligosaccharides in human serum. Glycobiology. 2014;24:542–53.
Tachibana K, Nakamura S, Wang H, Iwasaki H, Tachibana K, Maebara K, et al. Elucidation of binding specificity of Jacalin toward O-glycosylated peptides: quantitative analysis by frontal affinity chromatography. Glycobiology. 2006;16:46–53.
Saba J, Dutta S, Hemenway E, Viner R. Increasing the productivity of glycopeptides analysis by using higher-energy collision dissociation-accurate mass-product-dependent electron transfer dissociation. Int J Proteomics. 2012. doi:10.1155/2012/560391.
Trinidad JC, Barkan DT, Gulledge BF, Thalhammer A, Sali A, Schoepfer R, et al. Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol Cell Proteomics. 2012;11:215–29.
Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–62.
Baker PR, Trinidad JC, Chalkley RJ. Modification site localization scoring integrated into a search engine. Mol Cell Proteomics. 2011. doi:10.1074/mcp.M111.008078.
Baker PR, Chalkley RJ. MS-viewer: a Web-based spectral viewer for proteomics results. Mol Cell Proteomics. 2014;13:1392–6.
Hortin GL, Sviridov D, Anderson NL. High-abundance polypeptides of the human plasma proteome comprising the top 4 logs of polypeptide abundance. Clin Chem. 2008;54:1608–16.
Bern M, Kil YJ, Becker C. Byonic: advanced peptide and protein identification software. Curr Protoc Bioinformatics. 2012. doi:10.1002/0471250953.bi1320s40.
Catalina MI, Koeleman CA, Deelder AM, Wuhrer M. Electron transfer dissociation of N-glycopeptides: loss of the entire N-glycosylated asparagine side chain. Rapid Commun Mass Spectrom. 2007;21:1053–61.
Vakhrushev SY, Steentoft C, Vester-Christensen MB, Bennett EP, Clausen H, Levery SB. Enhanced mass spectrometric mapping of the human GalNAc-type O-glycoproteome with SimpleCells. Mol Cell Proteomics. 2013;12:932–44.
Bandeira N, Tsur D, Frank A, Pevzner PA. Protein identification by spectral networks analysis. Proc Natl Acad Sci U S A. 2007;104:6140–5.
Bandeira N. Spectral networks: a new approach to de novo discovery of protein sequences and posttranslational modifications. Biotechniques. 2007;42(687):689–91. passim.
Tabb DL, Vega-Montoto L, Rudnick PA, Variyath AM, Ham AJ, Bunk DM, et al. Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J Proteome Res. 2010;9:761–76.
Brennan SO, Myles T, Peach RJ, Donaldson D, George PM. Albumin Redhill (−1 Arg, 320 Ala–Thr): a glycoprotein variant of human serum albumin whose precursor has an aberrant signal peptidase cleavage site. Proc Natl Acad Sci U S A. 1990;87:26–30.
Peach RJ, Brennan SO. Structural characterization of a glycoprotein variant of human serum albumin: albumin Casebrook (494 Asp–Asn). Biochim Biophys Acta. 1991;1097:49–54.
Halim A, Brinkmalm G, Rüetschi U, Westman-Brinkmalm A, Portelius E, Zetterberg H, et al. Site-specific characterization of threonine, serine, and tyrosine glycosylations of amyloid precursor protein/amyloid beta-peptides in human cerebrospinal fluid. Proc Natl Acad Sci U S A. 2011;108:11848–53.
Steentoft C, Vakhrushev SY, Vester-Christensen MB, Schjoldager KT, Kong Y, Bennett EP, et al. Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat Methods. 2011;8:977–82.
Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32:1478–88.
Biemann K. Appendix 5. Nomenclature for peptide fragment ions (positive ions). Methods Enzymol. 1990;193:886–7.
Domon B, Costello CE. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj J. 1988;5:397–409.
Acknowledgments
The authors are grateful to Jonathan Trinidad and Jason Maynard for their useful advice on the WGA immobilization. We would like to thank Agnes Arva for the technical assistance. This work was supported by the following grants: Hungarian Scientific Research Fund 105611 (to Z. Darula); National Research, Development and Innovation Fund BAROSS-DA07-DA-ESZK-07-2008-0036 (to the Biological Research Centre, HAS, director P. Ormos); and New Szechenyi Plan GOP-1.1.1-11-2012-0452.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Published in the topical collection Glycomics, Glycoproteomics and Allied Topics with guest editors Yehia Mechref and David Muddiman.
Rights and permissions
About this article
Cite this article
Pap, A., Medzihradszky, K.F. & Darula, Z. Using “spectral families” to assess the reproducibility of glycopeptide enrichment: human serum O-glycosylation revisited. Anal Bioanal Chem 409, 539–550 (2017). https://doi.org/10.1007/s00216-016-9960-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-016-9960-7