Abstract
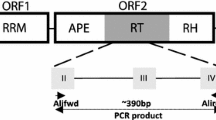
Retroviralenvelope (env)-like sequences in 2 cultivated allotetraploid cottons and their diploid progenitors have been identified and characterized in this study. DNA sequence analysis reveals that these sequences are heterogeneous. The observed sequence diversity, however, seems to preserve coding information. This is evidenced by the detection of the transmembrane domain (TM), which is the most conserved feature of the divergent retroviral env genes. The high ratio of synonymous to nonsynonymous changes suggests that these sequences are evolving under purifying selection. Phylogenetic analysis shows thatGossypium sequences closely cluster with a lineage of plant endogenous retroviruses that have anenv-like gene. These results provide evidence for the antiquity and the wide diversity ofenv-like sequences in theGossypium genome.
Similar content being viewed by others
References
Abdel Ghany AA, Zaki EA, 2002. Cloning and sequencing of an envelope-like gene inGossypium. Planta 216: 351–353.
Abdel Ghany AA, Zaki EA, 2003. Sequence heterogeneity of the envelope-like domain in the Egyptian cottonGossypium barbadense. Afr J Biotech 2: 341–44.
Abdel Ghany AA, Zaki EA, 2005. Isolation, characterization, and phylogenetic analysis of Bagy-2 retrotransposon envelope-domain in the Egyptian cottonG. barbadense. A J Biotech 8: 115–126.
Boeke JD, Eickbush TH, Sandmeyer SB, Voytas DF, 2000. Metaviridae. In: Murphy FA, ed. Virus taxonomy: ICTV VIIth Report, Springer, New York, 124–135.
Capy P, 2005. Classification and nomenclature of retrotransposable elements. Cytogenet Genome Res 110: 457–461.
Casacuberta JM, Santiago N, 2003. Plant LTR-retro-transposons and MITEs: control of transposition and impact on the evolution of plant genes and genomes. Gene 311: 1–11.
Chavanne F, Zhang DX, Liaud MF, Cerff R, 1998. Structure and evolution of Ty3/Gypsy family highly amplified in pea and other legume species. Plant Mol Biol 37: 363–375.
Eickbush TH, Malik HS, 2002. Origins and evolution of retrotransposons. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, eds. Mobile DNA II. ASM Press, Washington, D.C: 1111–1144.
Grandbastien MA, Lucas H, Morel JB, Mhiri C, Vernhettes S, Casacuberta TM, 1997. The expression of the tobacco Tnt1 retrotransposon is linked to plant defense responses. Genetica 100: 241–52.
Hawkins JS, Kim H, Nason JD, Wing RA, Wendel JF, 2006. Differential lineage-specific amplification of transposable elements is responsible for genome size variation inGossypium. Genome Res 16: 1252–1261.
Hirokawa T, Boon-Chieng S, Mitaku S, 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14: 378–79.
Hofmann K, Stoffel W, 1993. TMbase, a database of membrane spanning protein segments. Biol Chem 347: 166.
Hull R, 2001. Classifying reverse transcribing elements: a proposal and a challenge to the ICTV. Arch Virol 146: 2255–2261.
Kim A, Terzian C, Santamaria P, Pelisson A, Purd’ Homme N, Bucheton A, 1994. Retroviruses in invertebrates: thegypsy retrotransposon is apparently an infectious retrovirus ofDrosophila melanogaster. Proc Natl Acad Sci USA 91: 1285–1289.
Kim JK, Battini J-L, Manel N, Sitbon M, 2004. Emergence of vertebrate retroviruses and envelope capture. Virology 318: 183–191.
Kumar A, Bennetzen JL, 1999. Plant retrotransposons. Annu Rev Genet 33: 479–532.
Kumar S, Tamura K, Nei M, 2004. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Briefings in Bioinformatics 5: 150–163.
Laten HM, Majumdar A, Gaucher EA, 1998. SIRE-1, a copia/Ty1-like retroelement from soybean, encodes a retroviral envelope-like protein. Proc Natl Acad Sci USA 95: 6897–6902.
Laten HM, 1999. Phylogenetic evidence for Ty1-copia-like endogenous retroviruses in plant genomes. Genetica 107: 87–93.
Laten HM, Havecker ER, Farmer LM, Voytas DF, 2003. SIRE-1, an endogenous family fromGlycine max, is highly homogenous and evolutionary young. Mol Biol Evol 20: 1–13.
Lerat E, Capy E, 1999. Retrotransposons and retroviruses: analysis of theenvelope gene. Mol Biol Evol 16: 1198–1207.
Malik HS, Henikoff S, Eickbush TH, 2000. Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res 10: 1307–1318.
Misseri Y, Labesse G, Bucheton A, Terzian C, 2003. Comparative sequence analysis and predications for theenvelope glycoproteins of insect retroviruses. Trends Microbiol 11: 253–256.
Misseri Y, Cerutti M, Devauchelle G, Bucheton A, Terzian C, 2004. Analysis of theDrosophila gypsy endogenous retrovirusenvelope glycoprotein. J Gen. Virol 85: 3325–3331.
Nei M, Gojobori T, 1986. Simple methods forestimating the numbers of synonymous and nonsynonymous nucleotidesubstitutions. Mol Biol Evol3: 418–426.
Neumann P, Pozakova D, Koblizkova A, Macas J, 2005. Pigy, a new plant envelope-class LTR retrotransposon. Mol Gen Genomics 273: 43–53.
Pearce SR, 2007. SIRE-1, a putative plant retrovirus is closely related to a legume Ty1-copia retrotransposon family. Cell Mol Biol Lett 12: 120–126.
Pereira V, 2004. Insertion bias and purifying selection of retrotransposons in theArabidopsis thaliana genome. Genome Biol 5: 79–89.
Peterson-Burch BD, Wright DA, Laten HM, Voytas DF 2000. Retroviruses in plants? TIG. 16: 151–152.
Rost B, Casadio R, Farselli P, Sander C, 1995. Transmembrane helices predicted at 95% accuracy. Protein Sci 4: 521–533.
Saitou N, Nei M, 1987. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425.
Senchina DS, Alvarez I, Cronn RC, Liu B, Rong J, Noyes RD, et al. 2003. Rate variation among nuclear genes and the age of polyploidy inGossypium. Mol Biol Evol 20: 633–643.
Song SU, Gerasimova M, Kurkulos M, Boeke JD, Corces VC, 1994. An env-like protein encoded by aDrosophila retroelement: evidence thatgypsy is an infectious retrovirus. Genes Dev 8: 2046–2057.
Thompson JD, Higgins DG, Gibson TJ, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–6480.
Vanderwiel PS, Voytas DF, Wendel JF, 1993. Copia-like retrotransposable element evolution in diploid and polyploid cotton (Gossypium L.). J Mol Evol 36: 429–447.
Vicient CM, Kalendar R, Schulman AH, 2001. Envelope-class retrovirus-like elements are widespread, transcribed and spliced, and insertionally polymorphic in plants. Genome Res 11: 2041–2049.
Vitte C, Panaud Q, 2005. LTR retrotransposons and flowering plant genome size: emergence of the increase/decrease model. Cytogenet. Genome Res. 110: 91–107.
Voytas DF, 1992.Arabidopsis and cotton (Gossypium) as models for studying copia-like retrotransposons evolution Genetica 86: 13–20.
Wendel JF, Cronn R, 2003. Polyploidy and the evolutionary history of cotton. Advances in Agronomy 78: 139–186.
Wilhelm M, Wilhelm FX, 2001. Reverse transcription of retroviruses and LTR retrotransposons. Cell Mol Life Sci 58: 1246–1262.
Wright DA, Voytas DF, 2001. Athila4 ofArabidopsis and Calypso of soybean define a lineage of endogenous plant retroviruses. Genome Res 12: 122–131.
Wright DA, Voytas DF, 1998. Potential retroviruses in plants: Tat1 is related to a group ofArabidopsis thaliana Ty3/gypsy retrotransposons that encode envelope-like proteins. Genetics 149: 703–715.
Yano ST, Panbehi B, Das A, Laten MH, 2005. Diaspora, a large family of Ty3-gypsy retrotransposons inGlycine max, is an envelope-less memberof endogenous plant retrovirus lineage. BMC Evol Biol 5: 30–44.
Zaki EA, 2003. Plant retroviruses: structure, evolution and future applications. Afr J Biotech 2: 136–139.
Zaki EA, Abdel Ghany A, 2004. Ty3/gypsy retro-transposons in Egyptian cotton (G. barbadense). J C Science 8: 179–185.
Zaki EA, 2005. Ty1-copia group retrotransposon families in cultivated cottonsG. barbadense L. identified by reverse transcriptase domain analysis. DNA Seq 16: 288–294.
Zhang X, Wessler SR, 2004. Genome-widecomparative analysis of transposable elements in the related speciesArabidopsis thaliana andBrassica oleracea. Proc Natl Acad Sci USA 101: 5589–5594.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hafez, E.E., Abdel Ghany, A.A., Paterson, A.H. et al. Sequence heterogeneity of theenvelope-like domain in cultivated allotetraploidGossypium species and their diploid progenitors. J Appl Genet 50, 17–23 (2009). https://doi.org/10.1007/BF03195647
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF03195647